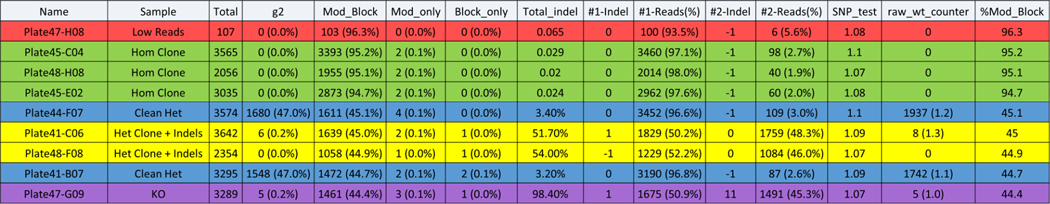
Figure 12: Analysis of CRIS.py data for screening single-cell clones.
Example of a .csv output file for screening single-cell derived clones for a point mutation project. The different clone types identified in the screen are labeled in the sample column. Clones with low read counts are highlighted in red. Homozygous clones (Hom Clone) are highlighted in green. Heterozygous clones (Clean Het) with the desired modification (Mod_Block) on one allele and the WT sequence on the other allele are highlighted in blue. Heterozygous clones with indels (Het Clone + Indels) on one allele and the desired modification (Mod_Block) on the other allele are shown in yellow. Clones with all out-of-frame indels (KO) are highlighted in purple. The top 2 indels are shown for representation purposes only.