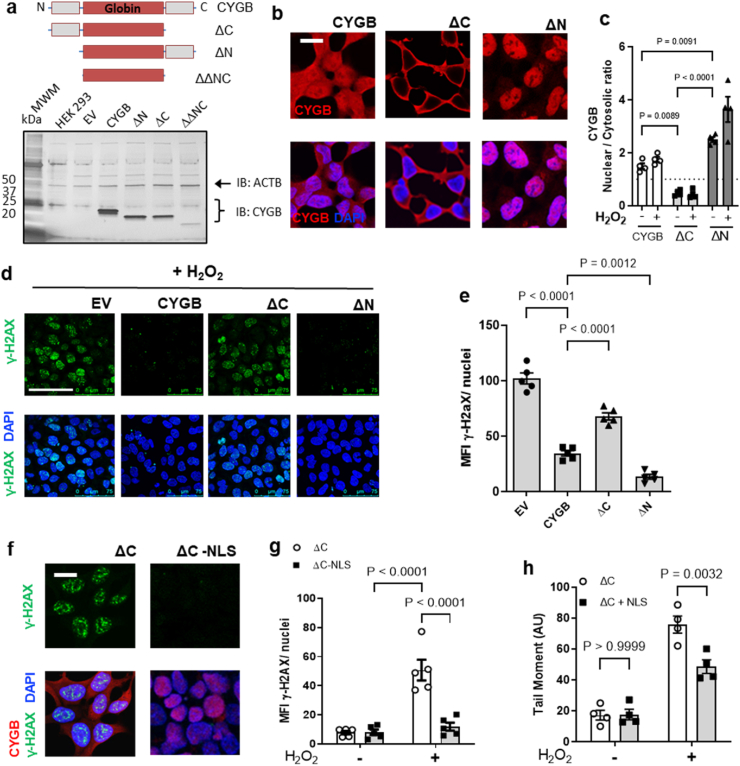
Fig. 4.
Nuclear cytoglobin is sufficient to inhibit DNA damage. (a) Top panel, schematic of the primary sequence of cytoglobin composed of a central globin domain and two unique 20 amino acid C- and N-terminal ends. Cytoglobin expression plasmids were generated for full length cytoglobin, and the N and C terminal truncated variants. Expression from the different plasmid in HEK293 was examined by Western blot. The double truncated variant did not express consistently. ACTAB is shown as a loading control. (b) Indirect immunofluorescence staining using an antibody directed against cytoglobin (red). (c) Quantitative analysis of the ratio of nuclear to cytosolic cytoglobin (CYGB) pixel intensity in HEK293 with plasmids expressing cytoglobin or empty plasmid (pcDNA) and treated with hydrogen peroxide. (d) Representative immunofluorescence images of γ-H2AX (green) staining in HEK293 expressing cytoglobin or empty plasmid following treatment with 200 μM hydrogen peroxide. (e) Quantitation of the mean fluorescence intensity of γ-H2AX per nuclei following 200 μM hydrogen peroxide treatment. (f) Representative immunofluorescence images of HEK293 cells expressing cytoglobin with C-terminal truncation and C-terminal truncation + nuclear localization sequence (NLS) exposed to 200 μM hydrogen peroxide and stained for γ-H2AX (green), DAPI (blue) and CYGB (red). (g) Quantitative analysis of γ-H2AX pixel intensity from results described in (f). Each point represents an independent experiment. (h) Quantitation of COMET tail moment from alkaline COMET assay in HEK C-terminal truncation and HEK C-terminal truncation + NLS exposed to 200 μM hydrogen peroxide. For statistical analysis throughout the figure, results were analyzed by one-way or two-way ANOVA followed by Tukey's post hoc test for multiple comparisons. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)