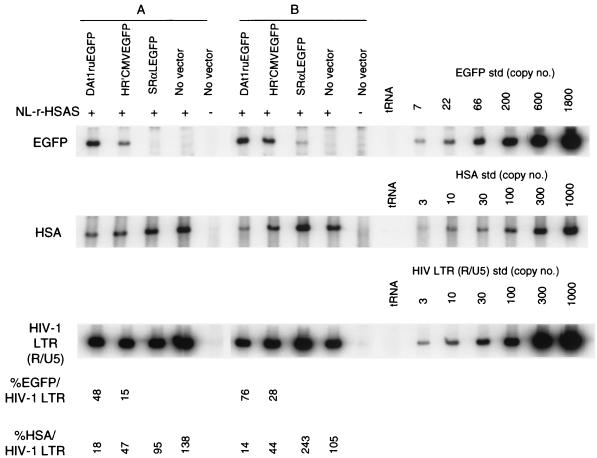
FIG. 3.
Quantitative analysis of de novo-synthesized vector and viral DNA from cells harboring both replication-competent HIV-1 and vectors. Cell culture supernatants were harvested from cultures of vector-transduced CEMx174 cells infected with NL-r-HSAS at day 4 (MOI of 1 [sample A]) and day 7 (MOI of 0.1 [sample B]). The p24 values of each of the supernatants of sample A were as follows: 1,180 ng/ml for the NL-r-HSAS-infected DAt1ruEGFP-transduced cells, 1,950 ng/ml for NL-r-HSAS-infected HR′CMVEGFP-transduced cells, 1,120 ng/ml for NL-r-HSAS-infected SRαLEGFP-transduced cells, 920 ng/ml for NL-r-HSAS-infected no-vector-transduced cells, and <0.08 ng/ml for mock-infected no-vector-transduced cells). The p24 values of each of the supernatants of sample B were as follows: 84.2 ng/ml for NL-r-HSAS-infected DAt1ruEGFP-transduced cells, 1,390 ng/ml for NL-r-HSAS-infected HR′CMVEGFP-transduced cells, 1,220 ng/ml for NL-r-HSAS-infected SRαLEGFP-transduced cells, 1,440 ng/ml for NL-r-HSAS-infected no-vector-transduced cells, and <0.08 ng/ml for mock-infected no-vector-transduced cells). Supernatants of sample B were normalized by the p24 value (84.2 μg/ml) for infection. Supernatants of sample A were used for infection without normalization. Supernatants were treated with DNase before infection, as described in Materials and Methods. Fresh CEMx174 cells (5 × 105) were infected for 2 h with 1 ml of each supernatant. At 12 h postinfection, DNA was purified from cells and subjected to quantitative PCR for EGFP gene, HSA gene, and HIV-1 R/U5 LTR sequences, as described in Materials and Methods. tRNA (0.1 μg/ml) was used as a negative control for PCR. Quantitative EGFP, HSA, and HIV-1 LTR (R/U5) DNA standards (std) were assayed in parallel. The EGFP- and HSA-specific signals were compared with that of the amplified HIV-1 LTR (R/U5) sequence to determine the percentages of EGFP/HIV-1 LTR and HSA/HIV-1 LTR, respectively. The data are representative of two independent PCR analyses.