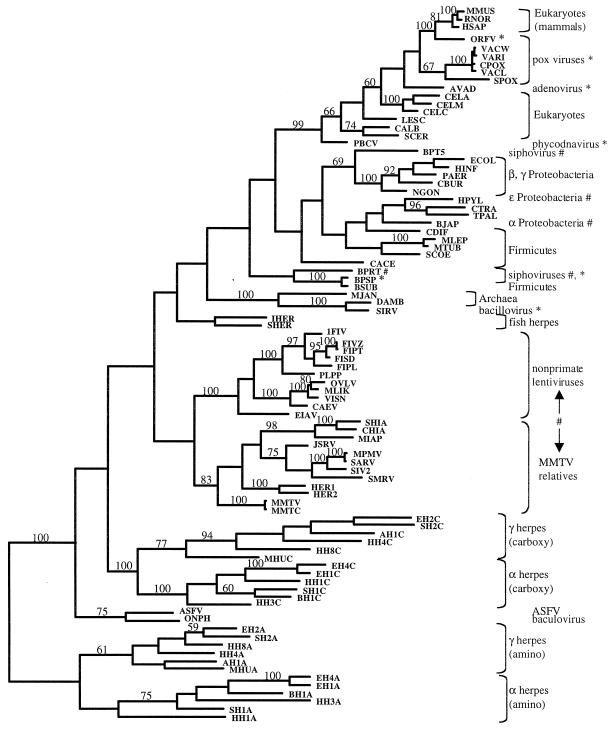
FIG. 4.
Shortest unrooted phylogenetic tree consistent with additive distances (Fitch-Margoliash algorithm). Poxvirus and avian adenovirus dUTPases are most similar to those of vertebrates. Chlorella virus (PBCV) dUTPase clusters with those of the eukaryotes. The bacteriophage r1t (BPRT) dUTPase clusters with those of the firmicutes. dUTPase from bacteriophage SPβ (BPSP) clusters with the sequence from host, B. subtilis (BSUB). Bacteriophage T5 (BPT5) also has a dUTPase similar to that of its proteobacterial host, E. coli (ECOL). Archaeal virus SIRV encodes a dUTPase similar to that of D. ambivalens (DAMB). Two proteobacterial dUTPases H. pylori (HPYL) and B. japonicum (BJAP) cluster with those of the groups Firmicutes, Chlamydiales, and Spirochaetales. MMTV relatives and nonprimate lentiviruses encode similar dUTPases. Strong cases for horizontal transfer supported by high bootstraps are indicated in this tree by an asterisk (∗). Other potential but less-well-supported cases are indicated with a pound symbol (#). Branch lengths are roughly proportional to distances, with the exception of very short branches, which are exaggerated for visibility. Groups of taxa corresponding to those listed in Table 1 are indicated with brackets and labels. Bootstrap values of 50% and higher are shown above supported branches. All taxon identifiers are listed in the Appendix.