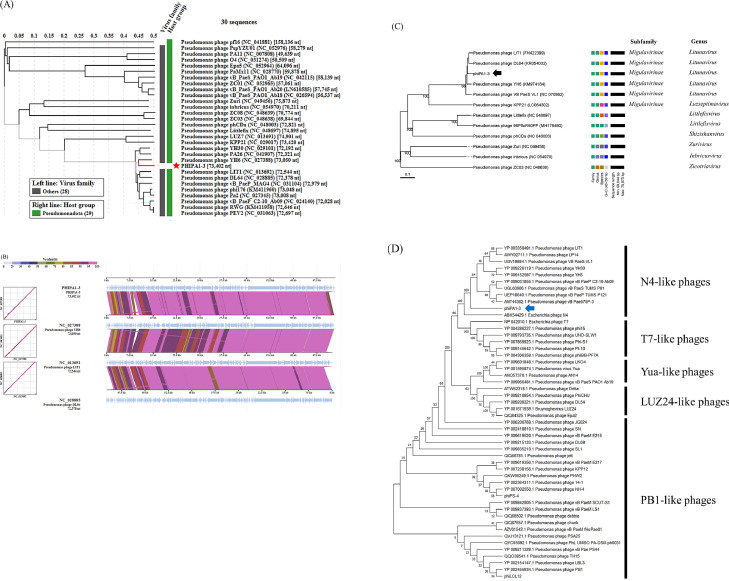
Fig. 7.
Genome comparison and phylogenetic tree analysis of phiPA1-3 with related phages. (A) The tree was constructed using VIPTree, by uploading phiPA1-3 genome sequence, and compared with all references in the database, phiPA1-3 was clustered with related Pseudomonas phages, the selection filter in Host group set with “Pseudomonadota”. (B) Alignment of selected genomes similar to phiPA1-3. Colored blocks show% identity of sequence. The comparison was carried out using VIPTree (https://www.genome.jp/viptree/). (C) The phylogenetic tree generated by VICTOR using the complete genomic sequence selected from various members of subfamilies and genera of Pseudomonas N4-like phages. (D) Phylogenetic tree of phage-encoded large terminase subunit (ORF85) of phiPA1-3. The amino acid sequences of the large terminase subunits were compared using MEGA11; the tree was generated using the neighbor-joining method and 1000 bootstrap replicates.