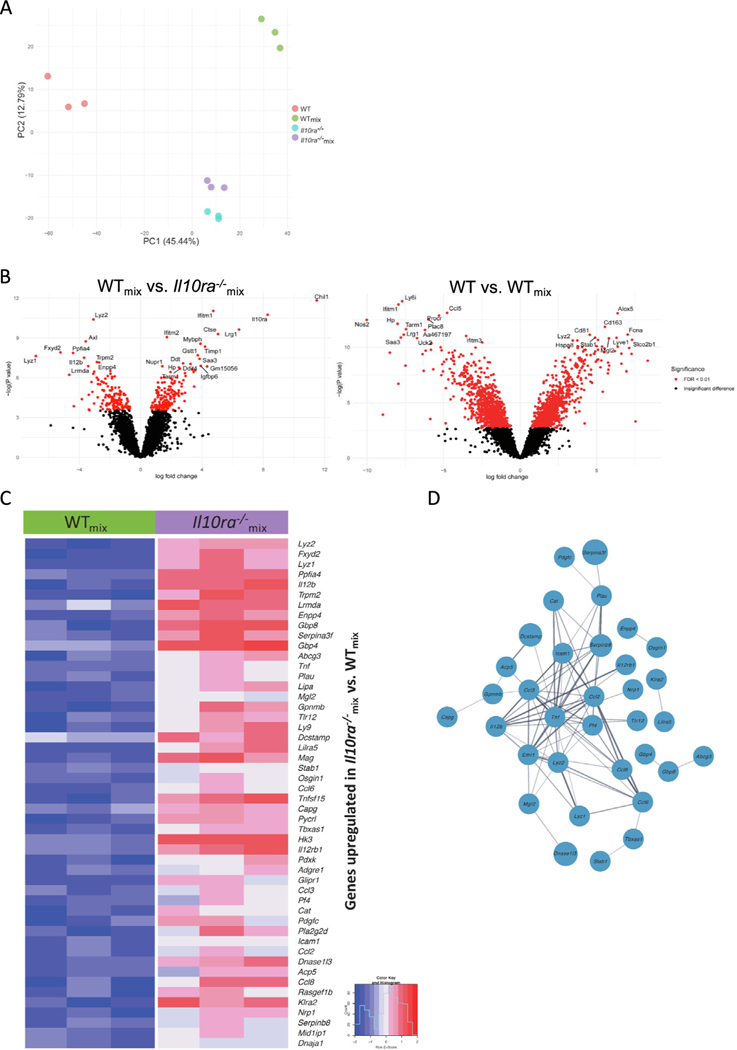
Fig. 6.
Bulk RNA sequencing from sorted colonic macrophages from mixed bone marrow chimeric mice. (A) Principal component analysis of sorted LP macrophages identifies cell autonomous and non-cell autonomous differences in gene expression caused by the absence of the IL-10R; (B) Left panel: comparison of WTmix versus Il10ra−/− identifies genes regulated in a cell autonomous fashion. Genes shown on the left arm of the volcano are expressed at lower levels in WTmix macrophages and those shown on the right arm of the volcano are expressed at higher levels in WTmix macrophages. Right panel: Comparison of WT versus WTmix reveals genes that are regulated in non-cell autonomous fashion by the presence of Il10ra−/− macrophages. Genes shown on the left arm of the volcano are expressed at lower levels in WT macrophages and those shown on the right arm of the volcano are expressed at higher levels in WT macrophages; (C) a heatmap depicts the expression of the top 50 upregulated genes in Il10ra−/− as compared with WTmix mice; (D) a network of physical and functional relations between the protein products of the top 50 upregulated genes in Il10ra−/−mix compared with WTmix mice, as determined by STRING v11.551, and visualized in Cytoscape 3.9.154. IL = interleukin; WT = wild-type.