Table 1.
Evolutionary rates estimates from root-to-tip regressions for the overall nucleotide changes, amino acid (aa) changes, and synonymous changes. The column ‘# of seq.’ shows the number of sequences that entered the analysis after filtering and restriction to the first 6 months after the emergence of the variant. The last three columns give the distances of the clade founder sequence from putative MRCA of SARS-CoV-2 (19B).
| Clade | # of seq. | Overall rate 
|
aa rate 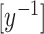
|
Syn rate 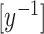
|
Overall div. | aa div. | Syn div. |
|---|---|---|---|---|---|---|---|
| 19B | 8,187 | 19.46 | 12.37 | 7.09 | 0 | 0 | 0 |
| 19B+ | 22,745 | 18.56 | 11.92 | 6.64 | 0 | 0 | 0 |
| 19B++ | 1,26,823 | 27.91 | 16.41 | 11.50 | 0 | 0 | 0 |
| 19A | 14,382 | 19.73 | 12.79 | 6.95 | 2 | 1 | 1 |
| 20A | 44,629 | 15.08 | 7.94 | 7.15 | 6 | 3 | 3 |
| 20B | 42,077 | 14.29 | 7.69 | 6.59 | 9 | 5 | 4 |
| 20C | 47,865 | 16.73 | 9.92 | 6.81 | 8 | 5 | 3 |
| 20A+ | 1,36,863 | 16.30 | 9.40 | 6.90 | 6 | 3 | 3 |
| 20E | 82,240 | 14.04 | 5.65 | 8.39 | 13 | 5 | 8 |
| 20H (Beta) | 5,077 | 15.48 | 9.51 | 5.96 | 21 | 16 | 5 |
| 20I (Alpha) | 2,77,226 | 11.56 | 5.49 | 6.07 | 30 | 19 | 11 |
| 20J (Gamma) | 24,970 | 12.97 | 6.29 | 6.68 | 35 | 22 | 13 |
| 21D (Eta) | 3,653 | 11.36 | 6.29 | 5.07 | 27 | 13 | 14 |
| 21G (Lambda) | 4,567 | 12.04 | 6.51 | 5.53 | 28 | 19 | 9 |
| 21H (Mu) | 2,897 | 18.25 | 10.05 | 8.20 | 31 | 21 | 10 |
| 21I (Delta) | 86,585 | 16.55 | 8.93 | 7.62 | 30 | 24 | 6 |
| 21J (Delta) | 7,33,825 | 14.30 | 7.17 | 7.14 | 34 | 28 | 6 |
| 21K (Omicron) | 9,56,696 | 13.48 | 6.45 | 7.03 | 56 | 46 | 10 |
| 21L (Omicron) | 6,90,177 | 9.24 | 4.39 | 4.85 | 68 | 50 | 18 |
| 22A (Omicron) | 33,451 | 7.71 | 2.81 | 4.90 | 71 | 52 | 19 |
| 22B (Omicron) | 1,24,239 | 11.90 | 5.03 | 6.87 | 66 | 50 | 16 |
