Figure 5.
mRNA-based longer RNA sensors.
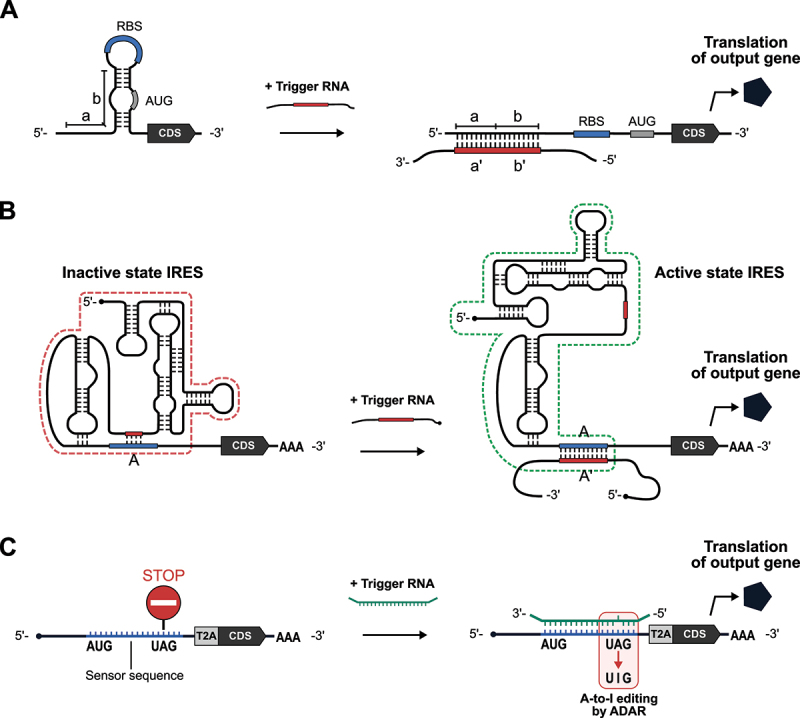
(A) Schematic illustration of bacterial toehold switches. In the absence of trigger RNA, ribosome binding site (RBS) and AUG initiation codon are sequestered. Interaction between a’ on the trigger RNA and a toehold sequence (a) on the toehold switch initiates RNA – RNA strand displacement interactions. The extended single-stranded sequence on the trigger RNA unwinds the hairpin structure through the b – b’ interaction, thereby initiating translation. (B) Schematic illustration of the eukaryotic version of toehold switch, eToehold. Short (red) and long (blue) RNA segments inserted into IRES (internal ribosome entry site) disrupt the functional structure of IRES. The trigger RNA – eToehold interaction (A – A’) restores the functional IRES structure and activates translation. (C) Schematic illustration of ADAR-mediated RNA sensors. In the absence of trigger RNA, translation terminates at the stop codon upstream of the CDS of an output gene. In the presence of trigger RNA, the RNA duplex surrounding the UAG stop codon recruits ADAR. ADAR-mediated A-to-I editing converts the UAG stop codon to the UIG codon for tryptophan, resulting in the translation of the downstream output CDS. The T2A peptide induces ribosome skipping to translate the output protein and the upstream CDS separately.
