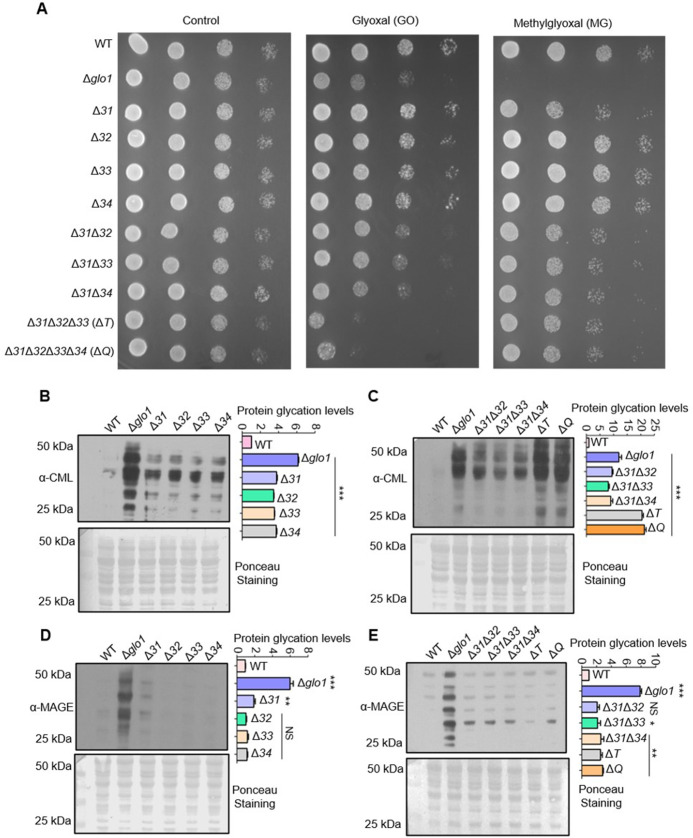
Figure 1. Deletion of Hsp31 paralogs induces protein glycation.
(A) Yeast phenotypic analysis. Cells were grown until the mid-log phase and harvested, subsequently treated with 10 mM MG before being spotted or spotted on YPD medium plates containing 15 mM GO. The plates were incubated at 30 °C and imaged at 36 hr. (B, C) Proteome glycation profile. Yeast strains were treated with 15 mM GO in the YPD culture medium and allowed to grow for 12 hr, followed by western analysis with anti-CML antibody. (D, E) MAGE detection by western blotting. Cells from the mid-log phase were incubated with 10 mM MG for 12 hr, and MAGE levels were estimated using an anti-MAGE antibody. Protein glycation levels were determined by measuring the whole lane intensities by densitometry and plotted with respect to WT. The blots stained with Ponceau S were used as the loading control. One-way ANOVA with Dunnet’s multiple comparisons test was used to determine significance from three independent biological replicates, *, p≤0.05; **, p≤0.01; ***, p≤0.001; NS, not significant.