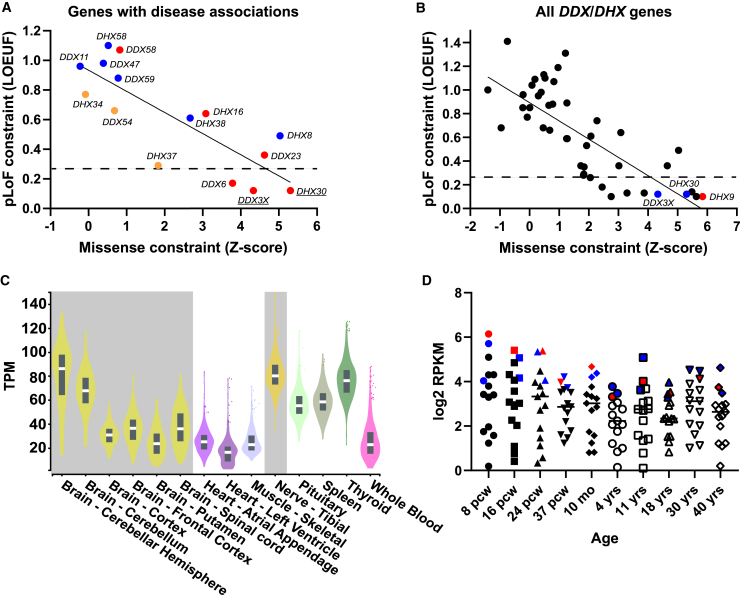
Figure 1.
Missense and LoF tolerance in the DDX/DHX superfamily and DHX9 expression
(A) Relationship between the pLoF and missense constraints among known or candidate DDX/DHX genes associated with disease phenotypes. Genes linked to dominant disease traits are shown in red, those linked to recessive traits are shown in blue, and those linked to mixed dominant and recessive traits are shown in orange. The solid line indicates linear regression. The dashed line shows the top LOEUF decile. The y axis shows LOEUF, and the x axis shows missense Z scores (gnomAD v2.1.1).
(B) Relationship between pLoF and missense constraint among all DDX/DHX genes. DHX9 is indicated by a red dot. Paralogs DHX30 and DDX3X are indicated by blue dots. The solid line indicates linear regression. The dashed line shows the top LOEUF decile. The y axis shows LOEUF, and the x axis shows missense Z scores (gnomAD v2.1.1).
(C) DHX9 mRNA expression in human adult tissues from the GTEx project. Nervous system tissues are highlighted in gray. The y axis shows transcripts per million (TPM).
(D) Average mRNA expression of known or candidate DDX/DHX genes associated with disease phenotypes in the developing nervous system from BrainSpan. DHX9 is indicated by red dots. Paralogs DHX30 and DDX3X are indicated by blue dots. The y axis shows log2 reads per kilobase million (RPKM), and the x axis shows the developmental stage. Abbreviations: pcw, post-conception weeks; mo, months; yrs, years.