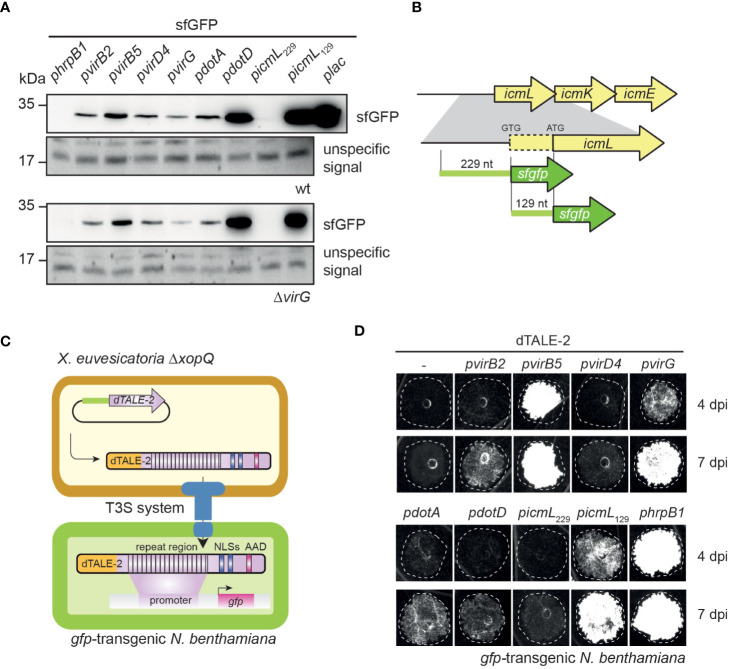
Figure 3.
The analysis of promoter-reporter fusions reveals in vitro and in planta activities of vir and icm/dot promoters. (A) vir and icm/dot promoters activate expression of the sfgfp reporter gene in X. euvesicatoria. Strains 85-10 and 85-10ΔvirG containing sfgfp under control of predicted promoters of virB2, virB5, virD4, virG, dotA, dotD and icmL on corresponding expression constructs were grown in NYG medium, and total cell extracts were analyzed by immunoblotting using a GFP-specific antibody. Expression constructs containing sfgfp under control of the hrpB1 promoter, which is inactive in NYG medium, and the lac promoter, which is active in X. euvesciatoria, were used as negative and positive controls, respectively. The experiment was performed three times with similar results. Unspecific signals detected by the GFP-specific antibody are shown to demonstrate equal loading. Given the high expression level of gfp in strain 85-10 containing gfp under control of the plac promoter, only 0.5 µl of the protein extract were loaded. (B) Schematic representation of icmL and predicted promoter regions. icmL is located upstream of icmK and icmE in the icm/dot T4S gene cluster on plasmid pXCV183 from X. euvesicatoria strain 85-10. A putative ATG start codon is located 129 nucleotides (nt) downstream of the annotated GTG translation initiation start site. For the analysis of putative icmL promoter regions, 229 nt upstream of the annotated start site and 129 nt upstream of the putative ATG start codon, respectively, were fused to sfgfp as indicated. (C) Principle of the dTALE-2-based in vivo expression and translocation assay. Expression constructs encoding dTALE-2 downstream of the analyzed vir or icm/dot promoter regions were introduced into X. euvesicatoria strain 85-10ΔxopQ which is pathogenic on N. benthamiana plants (Adlung et al., 2016). For translocation assays, bacteria were infiltrated into gfp-transgenic N. benthamiana plants which contain a viral vector construct encoding GFP under control of a dTALE-2-responsive promoter. Expression of dTALE-2 leads to its translocation by the T3S system because dTALE-2 contains an N-terminal secretion and translocation signal (depicted in orange). The central repeat region binds to specific bases in the dTALE-2-responsive promoter. C-terminal nuclear localization signals (NLSs) and an acidic activation domain (AAD) are required for nuclear localization and activation of in planta gene expression, respectively. Activation of gfp expression by dTALE-2 leads to GFP fluorescence in the infected leaf areas. (D) In vivo reporter assay for the detection of dTALE-2 translocation. X. euvesicatoria strain 85-10ΔxopQ with expression constructs containing dTALE-2 under control of predicted vir and icm/dot promoters as indicated was infiltrated into leaves of gfp-transgenic N. benthamiana plants containing gfp on a viral vector construct under control of a dTALE-2-responsive promoter. In planta activation of vir and icm/dot promoters allowed the synthesis and translocation of dTALE-2 via the T3S system, thus inducing GFP fluorescence which was monitored 4 and 7 dpi using a chemiluminescence/fluorescence imager which displays GFP fluorescence in black and white. The experiment was performed three times with similar results.