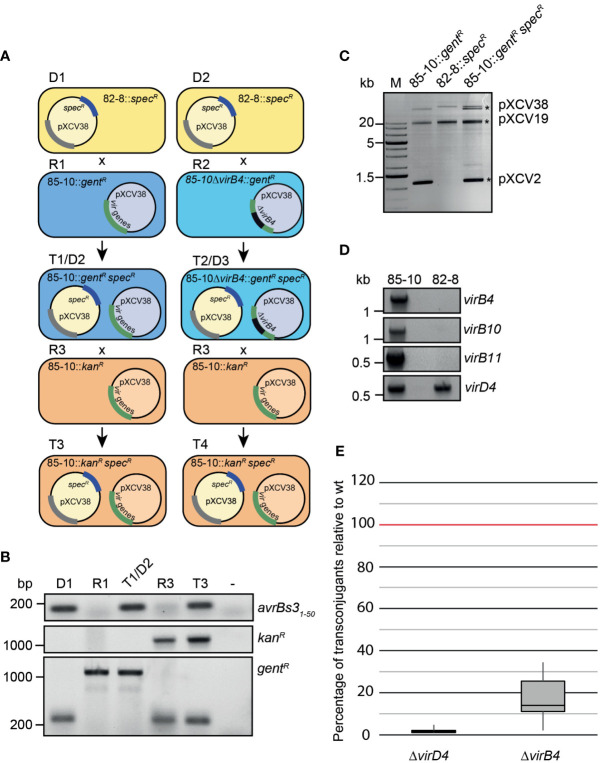
Figure 6.
Plasmid transfer by X. euvesicatoria strain 85-10 is controlled by VirD4. (A) Schematic overview on conjugation experiments with X. euvesicatoria strains. Plasmid pXCV38 from strain 82-8 (donor 1, D1) containing a spectinomycin resistance gene (pXCV3882-8::specR ) was transferred by conjugation to strain 85-10::gentR (recipient 1, R1) which contains the vir gene cluster on plasmid pXCV38. A grey bar indicates that the presence of a similar cluster on plasmid pXCV3882-8::specR is still unknown. The resulting transconjugant T1 (referred to as 85-10::gentR specR ) was gentamicin- and spectinomycin-resistant and contained both plasmids. The numbers of transconjugants from three independent experiments resulting from the transfer of plasmid pXCV3882-8::specR is given in Table S6 . Similarly, pXCV3882-8::specR was transferred to a derivative of strain 85-10::gentR with a deletion in virB4 (85-10ΔvirB4::gentR ; recipient 2, R2), resulting in transconjugants T2. T1 and T2 served as donor strains (D2 and D3) in a second conjugation experiment using strain 85-10::kanR , which contains a kanamycin resistance gene, as recipient (R3). The resulting transconjugants (T3 and T4) grew on medium with kanamycin and spectinomycin and contained plasmid pXCV3882-8::specR . In addition to the virB4 deletion mutant, we used virD4 mutant strains for the conjugation experiments (see panel E). virD4 is located on the chromosome and absent from plasmid pXCV38. (B) Confirmation of plasmid transfer by PCR analysis of transconjugants. The first 50 codons of avrBs3, the kanamycin and the gentamicin resistance gene were amplified by PCR using donor, recipient and transconjugant strains as depicted in (A) as templates. Amplicons were analyzed by agarose gel electrophoresis and visualized by an UV imager. Primers used to amplify the gentamicin resistance gene annealed to the chromosomal hpaFG region, thus leading to a smaller PCR product in the absence of the gentamicin resistance gene. (C) Analysis of isolated plasmids from X. euvesicatoria by agarose gel electrophoresis. Plasmids were isolated from X. euvesicatoria strains 85-10::gentR , 82-8 pXCV3882-8::specR (82-8::specR ) and 85-10::gentR pXCV3882-8::specR (85-10::gentR specR ) and separated by agarose gel electrophoresis. Asterisks indicate the expected sizes of plasmids pXCV2 (1852 bp), pXCV19 (19146 bp) and pXCV38 (38116 bp) from X. euvesicatoria strain 85-10. Plasmid pXCV183 (182572 bp), which is also present in strain 85-10, presumably did not migrate into the agarose gel. (D) PCR amplification of vir genes from X. euvesicatoria strains 85-10 and 82-8. vir genes were amplified by PCR using specific primers for virB4, virB10, virB11 and virD4 from strain 85-10 and genomic DNA of strains 85-10 and 82-8 as template. PCR products were analyzed by agarose gel electrophoresis and visualized by an UV imager. (E) VirD4 and VirB4 are required for efficient plasmid transfer. Donor and recipient were mixed at a ratio of 1:1 and incubated overnight on a cellulose nitrate filter placed on solid NYG medium and covered with 1% water agar. Serial dilutions were plated on selective plates to count the transconjugants. The highest numbers of transconjugants were obtained when the T4S wild-type strain 85-10::gentR containing pXCV3882-8::specR was used as donor and were set to 100%. Reduced numbers of transconjugants obtained with strains 85-10ΔvirD4 and 85-10ΔvirB4 both containing pXCV3882-8::specR are depicted in a boxplot diagram. Values and standard deviations represent the percentage of transconjugants from six experiments with strain 85-10ΔvirD4 and from ten experiments with strain 85-10ΔvirB4.