Figure 1.
CRISPRi screening in prostate cell lines decodes regulatory SNPs from eQTL analyses
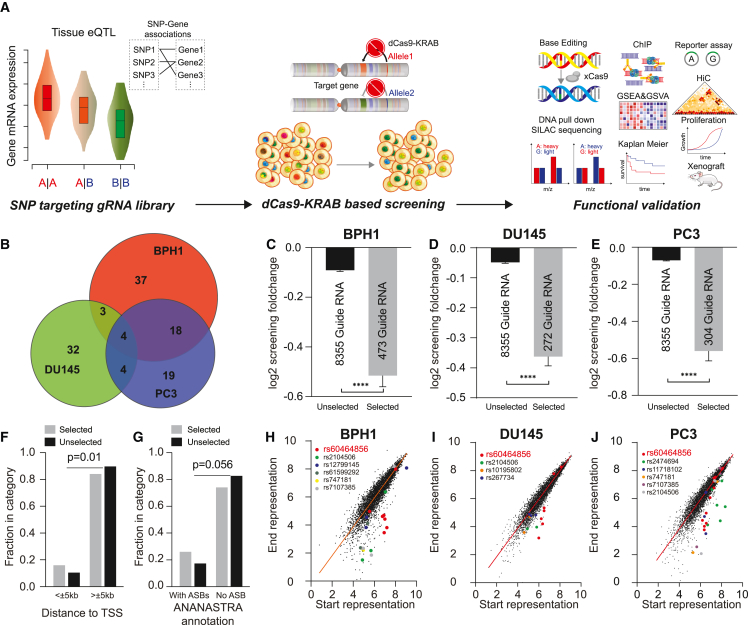
(A) Study design of the current project, including eQTL target selection, CRISPRi screening, and functional validation.
(B) Overall SNP candidates in BPH1 (red), DU145 (green), and PC3 (blue) cells over 3-week screening.
(C–E) Comparison of the gRNA fold change between the selected and unselected SNPs in BPH1 (C), DU145 (D), and PC3 (E) cells. The gRNA fold change was calculated from the average of the two biological replicates. The bar represents the standard error of the fold changes within each category.
(F) Enrichment of the selected compared to unselected SNPs in transcription start sites (TSSs).
(G) Enrichment of the selected compared to unselected SNPs in allele-specific binding (ASB) annotation according to the ANANASTRA database.
(H–J) Demonstration of gRNA representation for the selected SNPs in BPH1 (H), DU145 (I), and PC3 (J) cells. gRNAs targeting the main validation SNP rs60464856 are highlighted in red.