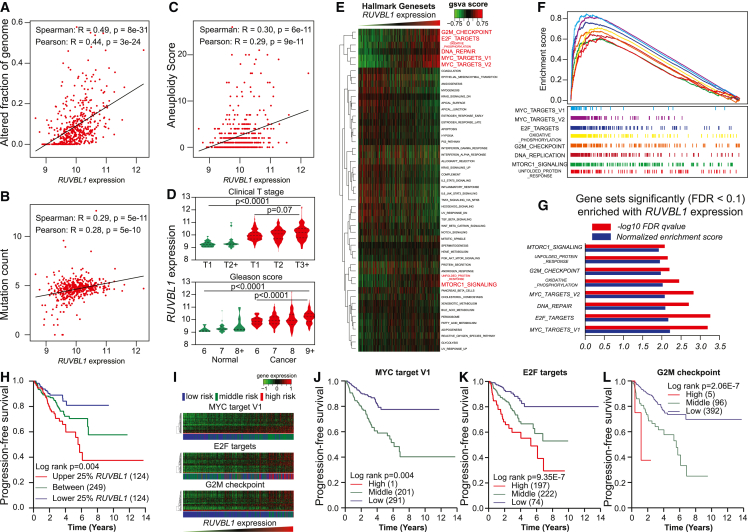
Figure 5.
Association between RUVBL1 expression and aggressiveness in prostate cancer
(A–C) Associations in TCGA prostate cancer cohort between RUVBL1 expression and genome instability metrics, including the altered fraction of genome (A), the mutation count (B), and the aneuploidy score (C).
(D) RUVBL1 expression in TCGA prostate tissue with different T stages and Gleason scores. The p value tested the linear trends between the RUVBL1 expression with the incremental stages or scores. The dashed lines indicate the quartiles and the medians of each group.
(E) GSVA on RNA-seq data in TCGA prostate cancer tissue by ranking RUVBL1 expression on HALLMARK collection pathways.
(F) GSEA enrichment plots showing significantly enriched HALLMARK pathways in TCGA prostate cohort.
(G) Normalized enrichment score and FDR q value for significantly enriched HALLMARK pathways in the TCGA prostate cohort.
(H) Kaplan-Meier analysis of progression-free survival in TCGA prostate cancer patients stratified by RUVBL1 gene expression.
(I) Heatmap with RUVBL1 leading-edge genes; the side bar shows k-means-stratified groups with different risks related to significantly enriched HALLMARK pathways, including MYC target V1, E2F targets, and G2M checkpoint, in the TCGA prostate cohort.
(J–L) Kaplan-Meier analysis on progression-free survival in TCGA prostate cancer patients stratified by the GSEA leading-edge gene enriched with RUVBL1 in each pathway, including MYC targets V1 (J), E2F targets (K), and G2M checkpoints (L).