Abstract
The mismatch repair protein MutS safeguards genomic integrity by finding and initiating repair of basepairing errors in DNA. Single-molecule studies show MutS diffusing on DNA, presumably scanning for mispaired/unpaired bases, and crystal structures show a characteristic “mismatch-recognition” complex with DNA enclosed within MutS and kinked at the site of error. But how MutS goes from scanning thousands of Watson-Crick basepairs to recognizing rare mismatches remains unanswered, largely because atomic-resolution data on the search process are lacking. Here, 10 μs all-atom molecular dynamics simulations of Thermus aquaticus MutS bound to homoduplex DNA and T-bulge DNA illuminate the structural dynamics underlying the search mechanism. MutS-DNA interactions constitute a multistep mechanism to check DNA over two helical turns for its 1) shape, through contacts with the sugar-phosphate backbone, 2) conformational flexibility, through bending/unbending engineered by large-scale motions of the clamp domain, and 3) local deformability, through basepair destabilizing contacts. Thus, MutS can localize a potential target by indirect readout due to lower energetic costs of bending mismatched DNA and identify a site that distorts easily due to weaker base stacking and pairing as a mismatch. The MutS signature Phe-X-Glu motif can then lock in the mismatch-recognition complex to initiate repair.
Significance
MutS scans the genome at high speed to find and initiate repair of basepair mismatches in DNA, thereby suppressing mutations and guarding against carcinogenesis. Structures of mismatch-bound MutS show DNA bent sharply and the mismatch site distorted within a “recognition” complex. But how does MutS check long stretches of DNA and form this recognition complex on encountering a mismatch? Microseconds-length molecular dynamics simulations provide the first atomic-resolution view of MutS actions in search mode. Both MutS and DNA undergo concerted conformational changes whereby MutS coarsely checks DNA shape and bendability for mismatch-induced differences and finely checks basepairs for deformability. Through this multipronged mechanism, MutS can quickly detect DNA segments that bend more easily and confirm sites therein that distort more easily as mismatches.
Introduction
How does a DNA repair protein find and fix infrequent but potentially disastrous basepairing errors in a vast background of canonical basepairs? The mismatch repair (MMR) pathway maintains genome integrity by correcting mistakes made by polymerases during DNA replication, including mismatched basepairs and nucleotide insertions/deletions, suppressing homeologous recombination, which involves erroneous basepairing between partially homologous DNA sequences and signaling cellular responses to damage lesions in DNA (1,2,3,4). MMR begins when MutS finds a basepairing defect in DNA through a process of search and recognition. This evolutionarily conserved protein forms a homodimer, e.g., Thermus aquaticus (Taq) or E. coli MutS (5,6), or heterodimer, e.g., S. cerevisiae or human Msh2-Msh6 (MutSα) or Msh2-Msh3 (MutSβ) (7,8), and uses its coupled, asymmetric mismatch binding and ATPase activities to initiate MMR (9). After recognizing a mismatch, MutS undergoes ATP binding-dependent conversion into a sliding clamp state (10,11,12,13) that can interact with MutL, an evolutionarily conserved protein that is itself an endonuclease (14) or interacts with an endonuclease, MutH, to nick the error-containing DNA strand (15,16). Subsequent controlled excision of the cut DNA strand past the mismatch site followed by accurate resynthesis completes MMR (17,18,19). Mutations in MutS and MutL proteins cause the hereditary Lynch cancer syndrome as well as several sporadic cancers, highlighting the importance of MMR in safeguarding the accuracy of genetic information (20,21).
In the past two decades, numerous crystal structures of MutS have been solved in complex with different mismatches and insertion/deletions—all showing the two subunits arranged in a closed, θ-shaped dimer with DNA enclosed in one chamber (Fig. 1; Taq MutS) (5,6,7,8,22). In this “mismatch-recognition” complex, DNA is sharply kinked at the mismatch site (45–60°), and a Phe and Glu residue from the mismatch binding domain (I) of MutS subunit A (equivalent to Msh6 in Msh2-Msh6) are stacked and hydrogen-bonded with the mispaired/unpaired base, respectively. Several other residues from the mismatch binding and clamp domains (IV) of both MutS subunits undergo hydrogen bonding and ionic interactions with the DNA backbone over about 5–8 nucleotides on either side of the mismatch. This large MutS footprint, encompassing more than one helical turn of DNA, and the observation that mismatch binding affinity and repair efficiency is influenced by neighboring nucleotide sequence (23,24), implies an important, although unclear, role for these nonspecific interactions during mismatch search and recognition. In addition to the structural data, single-molecule tracking of MutS proteins on DNA has provided a bird’s eye view of the search process (11,25,26). MutS encounters DNA via 3D diffusion, and it can encircle and move along the helix over thousands of basepairs at ∼700 bp/s via 1D rotational diffusion. A mismatch halts MutS movement, and it has been proposed that the conformational flexibility of DNA flanking the mismatch and its susceptibility to kinking enable MutS to stall and switch from scanning to mismatch-recognition state (27,28,29). But there is little atomic-resolution data to reveal the mechanism whereby MutS interrogates the flexibility of homoduplex DNA and identifies the odd basepairing error from the large excess of normal basepairs. An exciting breakthrough occurred recently when a cryo-EM structure of E. coli MutS bound to homoduplex DNA was solved (30). In this “scanning” complex, MutS clamp domains are separated, and the DNA is held in a straight conformation between the mismatch binding and clamp domains of the dimer (DNA-free MutS structures also indicate that clamp domains can adopt various conformations (5,13,31)). The stark differences between the scanning and mismatch-recognition complex structures attest to dynamic interactions and conformational changes in both MutS and DNA as the protein diffuses on the duplex in search of mismatches.
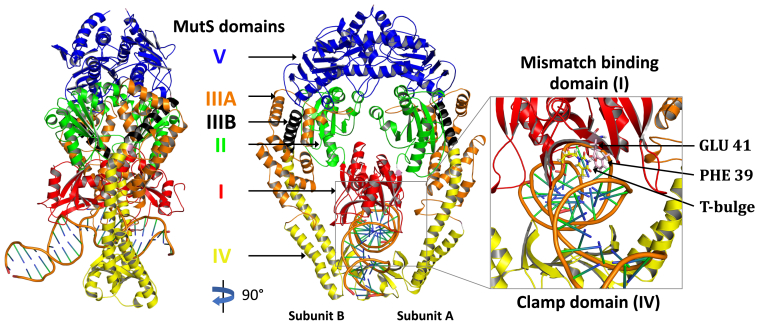
Figure 1.
Side and front view of the crystal structure of Taq MutS bound to mismatched DNA with a T-bulge (PDB: 1NNE). Each subunit in the MutS dimer comprises five domains: I (mismatch binding, red), II (connector, green), IIIA (lever, orange), IV (clamp, yellow), IIIB (lever clamp, black), and V (ATPase, blue). The mismatch detection interface formed by domains I and IV is highlighted in the inset (T-bulge as yellow sticks; mismatch binding residues Phe-39 and Glu-41 in subunit A as pink sticks).
Here, we report another substantive advance in elucidating the mismatch search and recognition mechanism employed by MutS on DNA. We conducted multiple independent 10–15 μs, unbiased molecular dynamics (MD) simulations of Taq MutS in complex with correctly basepaired homoduplex DNA and with DNA containing a single thymidine insertion (T-bulge). The data from these nonequilibrium dynamics simulations provide high-resolution temporal views of MutS interactions with the DNA backbone and bases spanning over almost two helical turns. Large conformational changes coupled through conserved contacts enable MutS to probe the helical shape, global bendability, and local basepair deformability of DNA, and transition from scanning mode to mismatch recognition at a bona fide error, such as a T-bulge, to initiate repair.
Materials and methods
Modeling MutS and DNA
The workflow of modeling MutS and DNA followed by MD simulations and analysis is outlined in Fig. S1. The Taq MutS protein sequence (811 residues) was retrieved from UniProt (32) (UniProt ID: Q56215). Of 811 residues, 765 could be accurately modeled using MutS structural data available in the RCSB databank (33). The crystal structure of Taq MutS-T-bulge (PDB: 1NNE) (34) was stripped of water molecules, and ADP-BeF3 was removed from both nucleotide binding sites. Residues 630–635 (loop) were missing in both Taq MutS subunits and were modeled on their counterparts in E. coli MutS (PDB: 2WTU) (35) using Modeller (36). ADP was modeled in subunit A using a related Taq MutS structure (PDB: 1FW6) (37). ADP was parameterized with Mg2+ coordinated by Ser-590, four water molecules, and an oxygen atom from the β-phosphorous using MCPB.py (38) methodology implemented in AMBER v18 (39) (Fig. S2). Gaussian (g16) (40) was used to estimate partial charges, bond distances, bond angles, torsions, and force constants for the ADP-Mg2+-water-serine complex. The calculations were performed using the B3LYP level of theory and 6-31G++ basis set in implicit water conditions. Mg2+-water bonds were treated using a nonbonded model with harmonic restraints while all other bonds were treated using the bonded model. The protonation state of amino acids was estimated using the H++ server (41). AMBER force field ff14SB (42) was utilized for amino acids and bsc0 (43) for DNA parameterization. Three additional DNA basepairs (G:C) were appended to the DNA to increase its length and modeled using Modeller. Taq MutS-homoduplex DNA was generated from MutS-T-bulge by removing the extra thymidine nucleotide in T-bulge DNA, followed by relaxation using Modeller and stepped minimization using AMBER v18 as described above. The t-LEaP module of AMBER v18 was used for hydrogen addition and general parametrization of amino acids and DNA basepairs.
MD simulations
MD simulations were performed on four constructs, two MutS-DNA complexes (10 μs each): 1) MutS-T-bulge in AADP-B0 liganded state and 2) MutS-homoduplex in AADP-B0 liganded state, and the two DNAs (15 μs each): 3) T-bulge and 4) homoduplex. Two additional independent 10 μs replicates of the MutS-homoduplex simulation were performed, starting with different initial velocities and atomic positions. MutS-DNA constructs were solvated in TIP3P (44) water molecules in a truncated octahedron box with 10 Å water clearance on each side. Na+ and Cl− ions were used to achieve electroneutrality and bring the system to 150 mM ionic strength. Periodic boundary conditions were employed along with the particle mesh Ewald algorithm (45,46,47) implementing a 10 Å cutoff to handle long-range electrostatic interactions. SHAKE (48) was used to restrict the motion of bonded hydrogen atoms; this also enabled use of 2 fs time steps in the simulations. Temperature and pressure were controlled using a Berendsen thermostat and barostat (49), respectively, with isotropic position scaling for pressure regulation. Random seeds were utilized to initiate the simulations. Minimization was achieved by a 5-step process (each comprising 2000 steps of steepest decent followed by 3000 steps of conjugate gradient), successively decreasing harmonic constraints on the MutS-DNA and DNA constructs in each of the 5 steps (100, 50, 20, 10, and 0 kcal/mol). Each construct was then heated to 300 K over 10 ps of simulation time. Stepped equilibration was performed in steps of 50 ps with successively decreasing harmonic constraints (100, 50, 10, 1, and 0.1 kcal/mol). Additional equilibration of 20 ns was performed without any restraints on MutS or DNA. Root mean-square deviation (RMSD), potential, kinetic, and total energy was monitored for structural and energetic stability during the equilibration. Production MD runs were carried out under NPT conditions. Trajectory frames were saved at intervals of 20 ps. All MD simulations were performed at Wesleyan University’s High Performance Computing Cluster using AMBER v18’s pmemd (50,51) implementation on nVIDIA GPUs.
Clustering, data visualization, and analysis
Dynamic cross-correlation matrix (DCCM) calculations, K-means clustering, and principal component analysis (PCA) were performed as implemented in the Cpptraj (52) module of AMBER. Clustering of the MutS-homoduplex trajectory data was performed based on RMSD. A shift analysis was performed by calculating the RMSD of each frame to the centroid of its assigned cluster and the centroids of all other clusters and shown as a frequency plot (53). The DNA substrates were analyzed using Curves+ (54). Incremental bending in the helical axis was determined at each basepair step in each frame of the trajectory and plotted as a function of simulation time. Images and videos were rendered using Gnuplot (55), VMD (56), PyMOL (57), and Chimera (58).
Results and discussion
Changes in MutS structural dynamics in the absence and presence of a mismatch
We performed several extended, unbiased MD simulations of two MutS-DNA complexes to compare the structural dynamics and interactions of MutS and DNA before and after mismatch detection. No high-resolution crystal structure of MutS bound to homoduplex DNA is available. Therefore, we remodeled DNA in the Taq MutS-T-bulge structure (PDB: 1NNE) by removing the extra unpaired thymidine in one strand to create homoduplex DNA. We also set the nucleotide-liganded state of the MutS dimer to AADP-B0. MutS exhibits asymmetric ATPase activity (only one subunit hydrolyzes ATP rapidly in the absence of a mismatch) (59,60,61), and the AADP-B0 state has been captured in many crystal structures of E. coli MutS bound to different mismatches in DNA (6,22). Both MutS-T-bulge and MutS-homoduplex complexes were simulated for 10 μs in explicit water conditions (note: two additional independent 10 μs simulations were performed for the MutS-homoduplex complex after analysis of the original trajectory revealed novel and dramatic conformational changes, described below). In addition, free T-bulge DNA and homoduplex DNA substrates in the absence of MutS were simulated for 15 μs. The workflow of modeling MutS and DNA followed by MD simulations and analysis is outlined in Fig. S1.
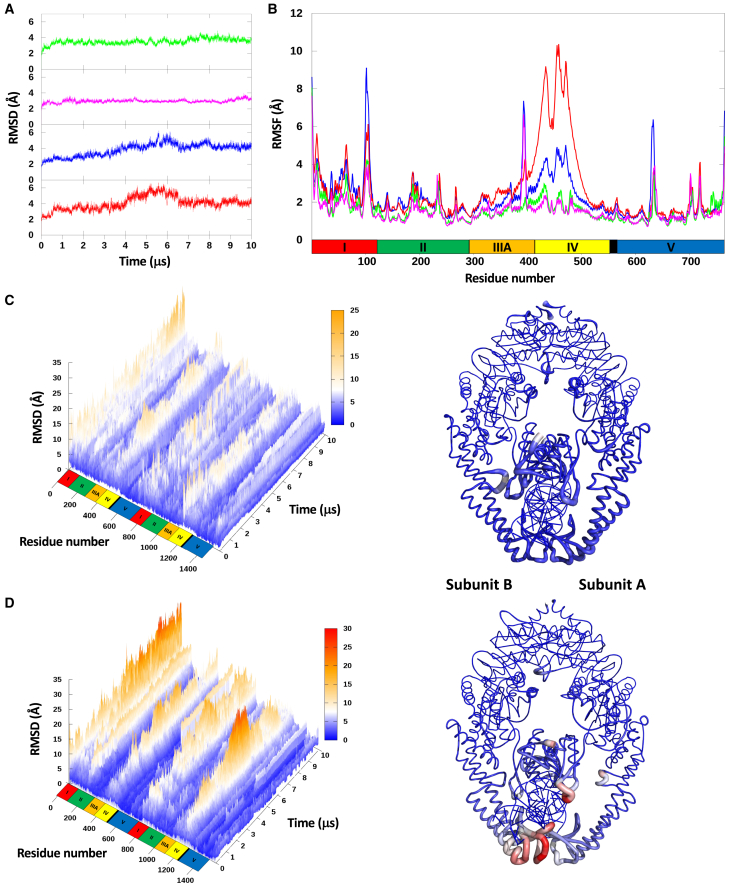
The RMSD values of each MutS subunit plotted over the entire simulation time show that MutS bound to T-bulge exhibits less conformational variability, with an average RMSD of 3.4 Å (0.35 SD) and 2.9 Å (0.18 SD) for subunits A and B in the T-bulge complex compared with 3.8 Å (0.8 SD) and 4 Å (0.86 SD) in the homoduplex complex, respectively (Fig. 2 A). This result is in accordance with the large number of crystal structures solved for MutS bound to mismatched DNA and none bound to homoduplex DNA. To resolve conformational variation at the amino acid level, we plotted the root mean-square fluctuation (RMSF) for each residue in the MutS subunits of both complexes (Fig. 2 B). Subunits A and B in the MutS-T-bulge complex did not diverge significantly from the initial structure, with RMSF values remaining at ∼2 Å for most residues (sharp spikes are due to loop regions). In contrast, both subunits in the MutS-homoduplex complex diverged significantly from the initial structure. The deviations were located primarily in clamp domain IV and were more pronounced in subunit B compared with A, revealing an asymmetric response in the MutS dimer to removal of the extra thymidine from DNA. At the other end of MutS, the RMSF values for the ATPase and dimerization domain V remained steady in all the subunits, consistent with the high stability of this domain observed in previous structural studies (5,62).
Figure 2.
RMSD, RMSF, and B-factor values from MD simulations of MutS in complex with T-bulge (MutS-T-bulge) or homoduplex DNA (MutS-homoduplex). (A) RMSD as a function of time and (B) RMSF for each residue in each subunit of MutS-T-bulge (subunit A, green; B, magenta) and MutS-homoduplex (subunit A, blue; B, red); MutS domains I–V and residue numbers are noted below the graph. (C) RMSD as a function of both residue number and time for MutS-T-bulge (left panel, variation from initial conformation: low [blue] to high [yellow]). B-factor variation of MutS-T-bulge superimposed on the initial structure (right panel; variation from initial conformation: low [blue/thinner line] to high [red/thicker line]). (D) RMSD as a function of both residue number and time for MutS-homoduplex (left panel; variation from initial conformation: low [blue] to high [red]). B-factor variation of MutS-homoduplex superimposed on the initial structure (right panel; variation from initial conformation: low [blue/thinner line] to high [red/thicker line]).
To assess the structural variation in MutS amino acids as a function of time, we aligned the MD trajectory ensembles with the ATPase domain and determined the RMSD for each residue during the simulation. There is little variation in MutS bound to T-bulge, with per-residue RMSD remaining under 10 Å through most of the simulation (Fig. 2 C, left). A B-factor plot of the trajectory relative to the initial conformation illustrates that the structure remains the same overall (Fig. 2 C, right). However, MutS dynamics are strikingly different when bound to homoduplex DNA, with large variations in per-residue RMSD occurring through most of the 10 μs simulation, often rising to 20–25 Å in parts of the protein (Fig. 2 D, left). The most notable structural variance occurs in the mismatch binding domain I of subunit A and clamp domain IV of subunit B, which make up the main “mismatch detection interface” and have the most extensive contacts with the unpaired thymidine and flanking nucleotides in the MutS-T-bulge complex (5) (Fig. 1). The B-factor plot illustrates the enhanced dynamics of these two domains when MutS is bound to homoduplex DNA (Fig. 2 D, right), in sharp contrast to when it is bound to T-bulge DNA (Fig. 2 C, right).
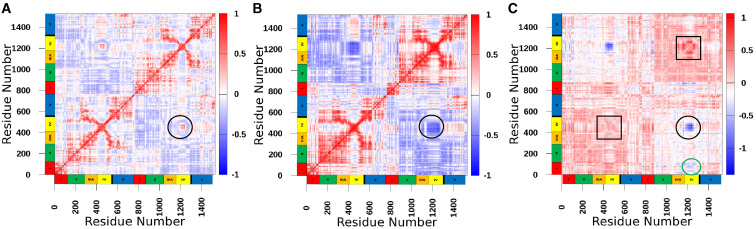
DCCMs were calculated for MutS in both complexes to examine the motions reported by the RMSD, RMSF, and B-factor values above (Fig. 3; summed over 10 μs). MutS bound to T-bulge displayed less motional correlation overall compared with MutS bound to homoduplex DNA (Fig. 3, A and B, respectively). The most prominent difference between the two complexes is in clamp domain IV, where positively correlated motions between subunits A and B in the MutS-T-bulge are negatively correlated in the MutS-homoduplex (black circle). These data indicate that the clamp domains move away from each other in the absence of a mismatch, which is consistent with reports of clamp separation in MD simulations of human MSH2-MSH6 in the absence of DNA (63) and in the recent cryo-EM structure of E. coli MutS bound to homoduplex DNA (30). The DCCMs plotted at 1 μs intervals revealed that the switch from positive to negative correlated motions in the MutS-homoduplex complex occurs at about 3 μs (consistent with RMSD data in Fig. 2 D), and remains negative through the end of the 10 μs simulation (Fig. S3). A difference plot of the DCCMs reveals more subtle details (Fig. 3 C). For instance, correlated motions in the subunit B clamp show greater deviation than in subunit A (black squares), indicating larger conformational changes in subunit B. Correlated motions between the subunit A mismatch binding domain I and subunit B clamp domain also differ between the two MutS complexes (Fig. 3 C, green circle), indicating concerted actions by the entire mismatch detection interface during the search and recognition process.
Figure 3.
Dynamic cross correlation matrices of MutS conformations summed over the MD simulation time (10 μs). The color scale ranges from red (high positive correlation) to blue (high negative correlation) in (A and B), and domains I–V and residue numbers of MutS subunits A and B are noted on the axis (left to right). (A) DCCM of MutS from the complex with T-bulge DNA. The black circle highlights a region of positive structural cross correlation between the clamp domains (IV) of subunits A and B. (B) DCCM of MutS from the complex with homoduplex DNA. The black circle highlights the same region, which now shows negative structural cross correlation between the clamps. (C) Difference plot between DCCMs of MutS from homoduplex and T-bulge complexes. The color scale ranges from red (high positive difference) to blue (high negative difference). Black squares highlight regions of higher deviation in subunit B clamp from the initial conformation compared with subunit A. The black circle highlights correlation between subunit A and B clamps. The green circle highlights correlation between subunit A mismatch binding domain (I) and subunit B clamp domain (IV) that occurs through contacts with DNA (see Fig. 6).
Changes in DNA structural dynamics in the absence and presence of a mismatch
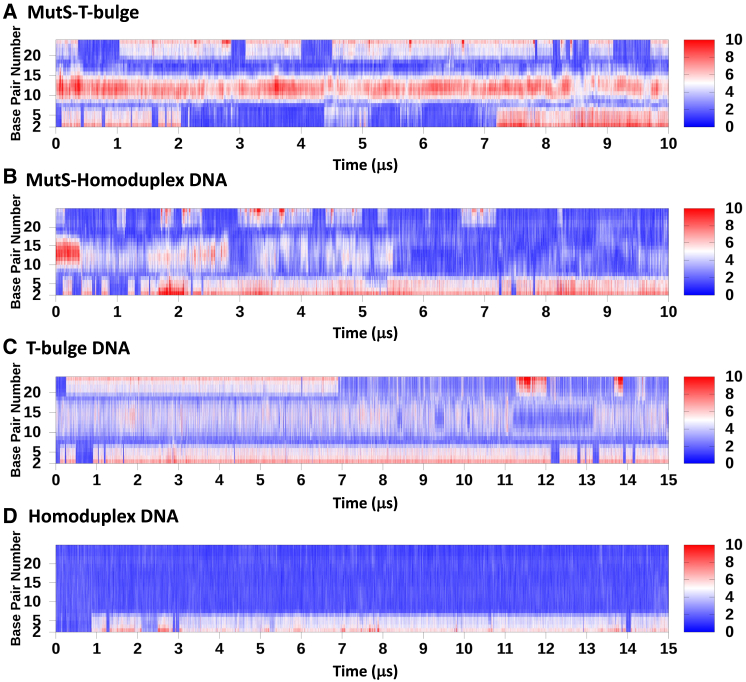
We also examined the role of DNA dynamics in differential interactions of MutS with homoduplex DNA versus mismatched DNA. The DNA substrates from MD simulations of both MutS-bound complexes were analyzed using Curves+ (54) to determine the degree of axial bending at each basepair step in each frame of the trajectory, which was plotted versus the simulation time (Fig. 4, A and B). The T-bulge and homoduplex DNA were also simulated independently of MutS for 15 μs and similarly analyzed to account for the effects of MutS on DNA dynamics (Fig. 4, C and D). MutS-bound T-bulge DNA was sharply kinked at the start and remained bent through the simulation, as expected from crystal structures (Fig. 4 A, middle red band in the trajectory; T-bulge at position 13); note: bending is elevated at the free DNA ends and exacerbated by an unpaired nucleotide at position 1 (PDB: 1NNE). In comparison, the free T-bulge DNA in the absence of MutS exhibited modest axial bending to various degrees through the 15 μs simulation (Fig. 4 C), in accordance with the conformational flexibility reported for DNA containing nucleotide insertions (64). The global bending propensity of this DNA in addition to local deformability at the T-bulge likely favors the kinked state stabilized by MutS in the mismatch-recognition complex (Figs. 1 and 4 A). This could explain the higher affinity of MutS for DNA containing an insertion compared with a mismatched basepair (65), which is not as intrinsically prone to bending (66,67).
Figure 4.
Axial DNA bending as a function of simulation time. Dynamic DNA bending at each basepair step was calculated in degrees (0–10°) using Curves+ software. The color scale ranges from red (high bend angle) to blue (low bend angle) and nucleotide numbers 1–23 are noted on the left (T-bulge at position 13). DNA bending in (A) MutS-T-bulge complex and (B) MutS-homoduplex complex. (C) T-bulge DNA and (D) homoduplex DNA bending in the absence of MutS. Note: elevated bending near the DNA end is because nucleotide 1 (G) is unpaired.
MutS-bound homoduplex DNA was bent by 45° in the starting complex formed by removal of the extra thymidine from T-bulge DNA in the crystal structure. This state lasted 0.48 μs, and then the homoduplex exhibited transient periods of unbending and bending between 0.48 and 5.5 μs (Fig. 4 B). After 5.5 μs, the homoduplex remained mostly unbent through to the end of the 10 μs simulation. In comparison, free homoduplex DNA in the absence of MutS did not display any axial bending and remained straight through the 15 μs simulation (Fig. 4 D). These results affirm that sharp kinking of homoduplex DNA is unfavorable to such an extent that it straightens out in the MutS-homoduplex complex despite structural hindrance from the protein in the initial conformation. As described in Figs. 2 and 3, MutS correspondingly converts to a more open state in which the clamp domains separate from each other. MutS-bound homoduplex DNA has been observed in straight and bent conformations by cryo-EM and atomic force microscopy (AFM), respectively (30,68,69). The MD trajectories reveal that these conformations are interconvertible, with DNA transitioning between straight-bent states and MutS between open-closed clamp states. These MutS-DNA dynamics may constitute a scanning mechanism for errors in DNA.
MutS and DNA conformational states on the path to finding a mismatch
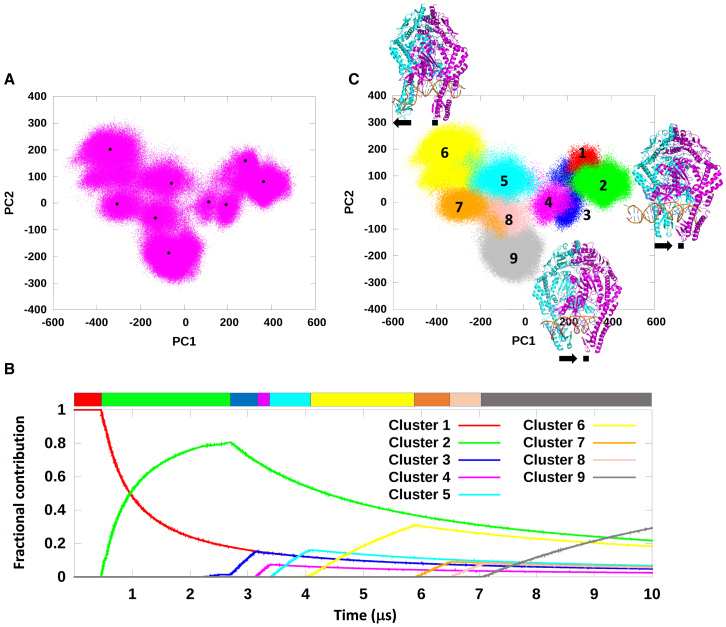
We analyzed the MutS-homoduplex trajectory data by principal component analysis and clustering (70) to address our hypothesis that the observed conformational dynamics constitute a mismatch scanning and detection mechanism. The top 10 principal components (PC) are shown in Fig. S4, and the first two components (PC1 and PC2) explain 70.64% of the conformational variation. PC1 mainly comprises motions of the clamp domains moving apart, with accompanying adjustment of the mismatch binding domains and translation of DNA (Video S1). PC2 mainly comprises DNA unbending motions (Video S2). Together these components account for the dominant changes in MutS-homoduplex during the simulation. Plotting the trajectory frames as a function of PC1 and PC2 parsed the data into nine different conformational groups (Fig. 5 A, black dots are the centroids). A shift analysis was performed to assess the separation between the nine clusters by calculating the RMSD of each frame to the centroid of its assigned cluster and the centroids of all other clusters. The data in Fig. S5 show that the frames in a given cluster lie closer to their own centroid and are distant from the centroids of the other clusters, which supports their classification as distinct conformational populations of the MutS-homoduplex complex. Further analysis of the full trajectory data by K-means clustering based on RMSD between conformations provided additional support that the nine clusters are structurally distinct from each other. Moreover, the fractional composition of each cluster plotted over time showed that they form and decay sequentially through the simulation (Fig. 5 B; clusters numbered 1–9 in order of appearance). Fig. 5 C shows the clustering data projected onto the PC1 and PC2 components, colored and numbered 1 through 9 by cluster as identified in Fig. 5 B. Together, the results indicate a kinetic pathway in which the MutS-homoduplex complex transitions between bent and straight DNA conformations through a series of different substates (Videos S3 and S4; detailed description provided below).
Figure 5.
Principal component and clustering analysis of the MutS-homoduplex DNA complex MD simulation. (A) MutS-homoduplex conformations projected as a function of the top 2 principal components of PCA (pink dots). The data sort into nine structurally distinct clusters (centered at the black dots). (B) Fractional composition of MutS-homoduplex clusters as a function of time. Clustering analysis was performed using K-means clustering based on RMSD. The kinetic trace for each cluster (substate) is numbered in the order of appearance and has a different color (substate 1, red; 2, green; 3, blue; 4, magenta; 5, cyan; 6, yellow; 7, orange; 8, pink; and 9, gray). (C) Individual substates from the clustering analysis projected as a function of PC1 and PC2. Conformations of prominent substates 2, 6, and 9 are shown (see Fig. 6 and Table S1). Similar results were obtained from two replicate MutS-homoduplex simulations, shown in Figs. S6 and S7.
The observed motions are primarily separation of MutS clamp domains (IV) and lateral movement of DNA.
The observed motions are primarily unbending of homoduplex DNA from kinked to straight conformation.
Key contact residues are shown (described in Fig. 7). The overall DNA shape, bendability, and local base stacking and pairing stability could be assessed through dynamic, concerted interactions of MutS with the sugar-phosphate backbone and bases over one to two helical turns.
A concern that these striking results are from a single, albeit long, simulation was addressed by performing two additional 10 μs MD simulations of the MutS-homoduplex complex, starting from different initial velocities and structures. The data were analyzed as described above and the results are shown in Figs. S6 and S7. These new simulations again reveal clustering of MutS-homoduplex into nine distinct conformational substates that transition in series along the trajectory. We also see evidence that substates 8 and 9 can transition to substates 3–4, suggesting that the complex can cycle through parts of the pathway. We also tested the significance of the transitions between substates individually by selecting representative structures from each one 1–8 and simulating them independently for at least 2 μs each. The trajectories were projected onto to the PC1 and PC2 components obtained from the original full 10 μs simulation (Fig. S8). The data confirm that each substate can transition to the next one in the pathway, and indicate that some steps maybe reversible (e.g., between substates 5 and 6).
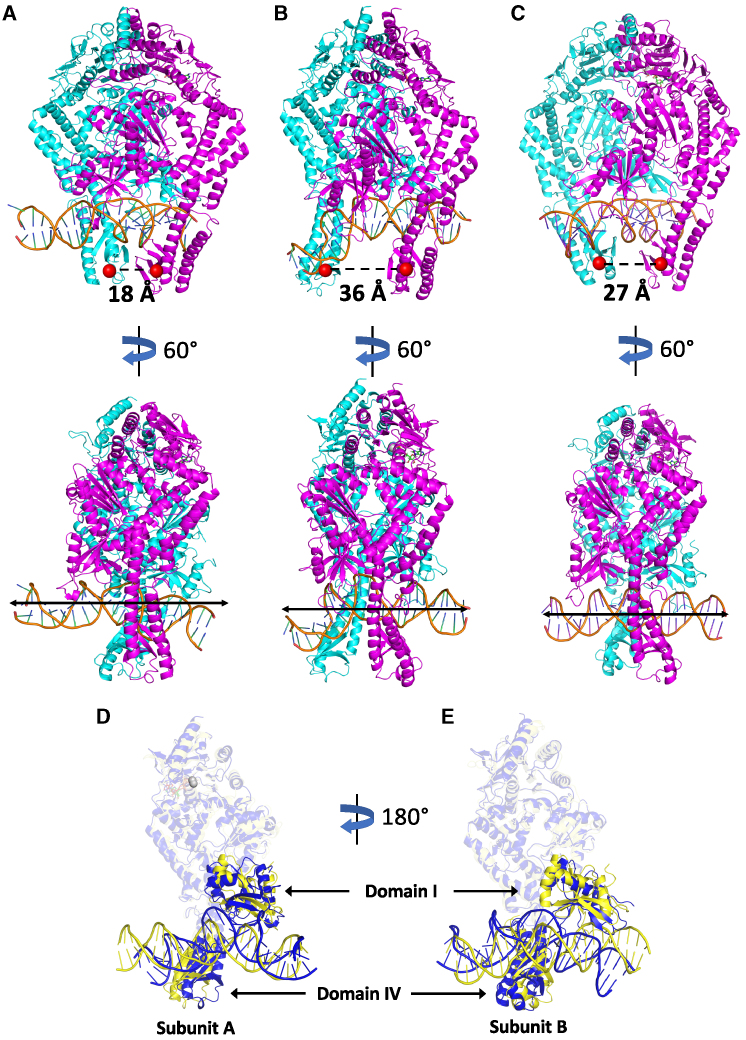
The nine MutS-homoduplex substates have different lifetimes, reflecting varying degrees of stability (Fig. 5 B and Table S1). Three substates have longer lifetimes than the others: 2 (2.18 μs), 6 (1.83 μs), and 9 (2.93 μs). Fig. 6 illustrates representative structures of these three prominent substates. Similar structures were obtained for substates 2, 6, and 9 from the two replicate MutS-homoduplex trajectories as well, further highlighting the significance of these conformational populations in MutS-homoduplex dynamics (Figs. S6 and S7). In substate 2, the DNA has almost straightened out from the initial bent state; however, there is little change in the overall MutS conformation and the clamp domains remain closed, reflecting the high stability of the dimeric θ structure (Fig. 6 A; clamps separated by 18 Å). In substate 6, the subunit B clamp has moved forward on DNA relative to subunit A, resulting in a separation of 36 Å between the two clamp domains (Fig. 6 B); Fig. S6 shows that this separation can be as wide as 50 Å. Notably, the DNA is bent, held out of plane through newly formed contacts with the subunit B clamp. In substate 9, the subunit B clamp has moved back closer to subunit A, although the two clamp domains remain partially open (separated by 27 Å) and the DNA is fully straight (Fig. 6 C). A coarse-grained simulation of human MSH2-MSH6 has shown that the protein does not dissociate from DNA even when the clamp domains are modeled in a relatively open position (71). This finding provides strong support for our proposition, discussed in detail below, that the open-to-closed conformational dynamics of clamp domains underlie the mismatch search mechanism employed by MutS as it diffuses on homoduplex DNA (E. coli MutS diffuses on DNA for ∼1 s per binding event (11)). In addition to clamp domains, the mismatch binding domains also undergo rearrangement in concert with the changes in DNA. An overlay of the initial and final complex conformations highlights the changes in the mismatch binding and clamp domains of subunit A (Fig. 6 D) and subunit B (Fig. 6 E) and the corresponding changes in homoduplex DNA.
Figure 6.
Prominent MutS-homoduplex DNA complex substates observed during MD simulation. Subunit A, magenta; subunit B, cyan (A–C); distance between clamp domains IV (dashed line). (A) Substate 2 (straightened DNA, clamps very close), (B) substate 6 (bent DNA, clamps farthest apart), and (C) substate 9 (straight DNA, clamps close). Corresponding 60° rotated views show DNA conformation. Similar substate 2, 6, and 9 conformations were obtained from the two replicate MutS-homoduplex simulations as well (Figs. S6 and S7). (D) Overlay of initial (blue) and final (yellow) subunit A conformations, showing rearrangement of the mismatch binding (I) and clamp (IV) domains and coupled DNA unbending (subunit B is hidden and the other subunit A domains are translucent). (E) 180° rotation shows the analogous view of subunit B.
Representative structures of all substates 1 through 9 can be viewed in series in Videos S3 and S4. Homoduplex DNA adopts a range of straight-to-bent conformations and MutS adopts open-to-closed clamp conformations in these substates. After DNA first straightens out in substate 2, subsequent movements evoke a cantilever, with MutS anchoring the DNA polymer through relatively stable contacts at one location with the mismatch binding and clamp domains of subunit A (magenta) and enabling it to bend (substate 6) and unbend (substate 9) through dynamic contacts at other locations with the mobile clamp domain of subunit B (cyan). The videos also provide a preview of interactions between MutS residues and DNA, whose role in interrogating DNA to detect a basepairing error is discussed in detail below.
MutS-DNA interactions assess DNA shape, global bending, and local basepair deformability in the search for mismatches
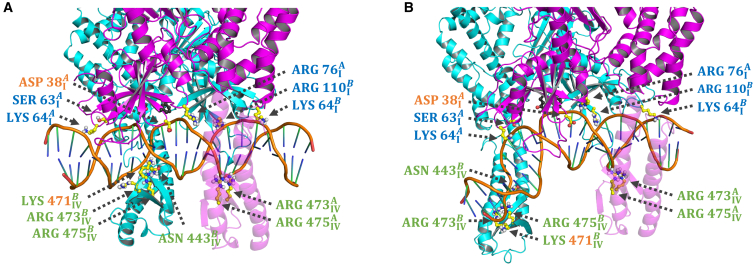
Since the first crystal structures of E. coli and Taq MutS encircling mismatched DNA were solved (5,6), it has been proposed that the protein employs an indirect readout mechanism, testing the conformational flexibility of DNA to find sites prone to bending and deformation. The conformational dynamics of Taq MutS-homoduplex DNA complex reported here provide, for the first time, a molecular underpinning for this hypothesis. Fig. 7 illustrates several interactions between MutS residues and homoduplex DNA in substate 9 (straight DNA) and substate 6 (bent DNA), selected on the basis of contact frequency (>20% of all frames in the substate trajectory have at least one hydrogen bond; Table S2). The interactions can be categorized into three functional classes. The first set of residues is distributed across the mismatch binding domains of both subunits and contacts the sugar-phosphate backbone in both substates 9 and 6 (Fig. 7, A and B, blue labels). Ser- and Lys- from subunit A and Lys- and Arg- from subunit B bookend about 1.5 helical turn and could check DNA shape as MutS undergoes 1D rotational diffusion along the axis in scanning mode (11). Arg- is an additional point of contact with the DNA backbone midway between the flanking Lys-64 residues. The second set of residues is concentrated in the clamp domains of both subunits and is involved in large conformational changes that could check the global bendability of DNA (Fig. 7, A and B, green labels; Videos S3 and S4). Arg- and Arg- from subunit A maintain contact with the same site on DNA through the entire MD trajectory (along with Arg- from the mismatch binding domain). In stark contrast, Asn-, Lys-, Arg-, Arg- from subunit B contact different sites on DNA when the clamp extends to a fully open conformation as the DNA bends (substate 6) or retracts to a partially closed conformation as the DNA straightens (substate 9); note: the highly conserved Lys- residue is also likely part of this set, but does not present as a high-frequency contact because the DNA is short. Extension and retraction of the subunit B clamp coupled with DNA bending and unbending, respectively, can be seen clearly in Videos S3 and S4 (substates 1–9 linked in series). Finally, at the center of the MutS footprint on DNA, Asp- remains inserted in the minor groove during the entire trajectory while Lys- is inserted in the corresponding major groove when the DNA is straight, as in substate 9 (Fig. 7, A and B, orange labels). These two highly conserved residues evoke a pincer that could check local deformability by destabilizing stacking and hydrogen bonding between bases (5,6,7).
Figure 7.
MutS-DNA interactions constituting the mismatch search mechanism. Three classes of interactions are shown for (A) substate 9 and (B) substate 6 from the MD trajectory of MutS-homoduplex DNA complex. Subunit A, magenta; subunit B, cyan; residues shown as ball-and-stick, yellow; subunit A Phe-39 and Glu-41 that lock in the mismatch-recognition complex at a bona fide error, black. Contacts from the mismatch binding (I) and clamp (IV) domains are shown that could enable MutS to check 1) the shape of DNA over ∼1.5 helical turn (blue labels), 2) the bendability of DNA (green labels) as subunit B clamp extends (substate 6) and retracts (substate 9) along the axis, and 3) the local deformability of basepairs at the center of the MutS binding site on DNA (orange labels).
For a Figure360 author presentation of this figure, see https://doi.org/10.1016/j.bpj.2023.06.006.
We noted that Phe-, which undergoes the signature π-π stacking with the mispaired/unpaired base in the mismatch-recognition complex, is twice as flexible in the MutS-homoduplex versus the MutS-T-bulge trajectory. This finding is consistent with previous AFM studies that showed similar bending of homoduplex DNA by Taq MutS wild-type and Phe-39 (Ala) mutant (69), suggesting that Phe-39 does not make significant contact with the homoduplex during the mismatch search process. On the other hand, almost all of the residues listed above that undergo nonspecific electrostatic and hydrogen bonding interactions with the homoduplex also interact with kinked T-bulge DNA in the mismatch-recognition complex, reflecting their comprehensive role in both search and recognition mechanisms. Moreover, these residues are also conserved in diverse organisms, including E. coli MutS and human, mouse, and/or S. cerevisiae Msh2-Msh6, affirming their importance in this fundamental DNA repair pathway (5,6,7). Structural alignment of the Taq MutS (PDB: 1NNE) and human MSH2-MSH6 (PDB: 2O8F) complexes with T-bulge DNA shows that the conserved residues are positioned to perform analogous functions in both proteins (Fig. S9 and Table S3). Several are also reported in the InSiGHT database of variants for Lynch syndrome, although the effects of the point mutations are unknown (72).
Together the dynamic interactions described above constitute an indirect readout mechanism that combines a coarse search for regions of unusual conformational flexibility in DNA and a fine search that discriminates between a canonical basepair and a mismatch (Videos S3 and S4). At a coarse level, MutS switches between conformations that enable DNA bending and unbending as it diffuses along the helix, and the lower energetic cost of bending a mismatch-containing segment could help localize the protein to a potential target site. These actions alone are insufficient as MD studies of DNA bending and deformability have shown that bending propensity contributes to mismatch detection but does not confer high enough selectivity (29,67), which is consistent with reports from AFM analysis of MutS-homoduplex DNA complexes as well (69). At a fine level, MutS interactions could preferentially promote destabilization and opening of a mismatched basepair and enable kinking of DNA. This mechanism is consistent with the report that basepair opening energetics account for the higher affinity of MutS for mismatched over homoduplex DNA (73). At this stage, the signature Phe- and Glu- residues from subunit A can stack and hydrogen bond with the displaced base and lock in the mismatch-recognition complex (Fig. 1).
Implications of asymmetry in MutS structure and function for mismatch repair
Despite being a homodimer, bacterial MutS is a functional heterodimer whose subunits have asymmetric mismatch recognition and ATPase activities (9). Our data reveal striking DNA binding asymmetry even in the mismatch search mechanism, with distinct roles for the two clamp domains in bending DNA. It is not known if the subunits originally designated as A and B in the homodimer (from the MutS-T-bulge complex) retain their roles while scanning homoduplex DNA, or if they can be switched. A recent study of mixed wild-type/mutant E. coli MutS dimers found that subunit functions are exchangeable for initiation of repair after mismatch recognition (30), which supports the possibility that switching occurs during the mismatch search phase as well. In eukaryotic heterodimers, DNA binding asymmetry is codified, with Msh6 and Msh2 serving as subunits A and B, respectively. The division of labor in the mismatch search mechanism employed by bacterial homodimers may have allowed Msh2 to retain the common function of checking global DNA bendability through evolution, while Msh6 and Msh3 specialized to check local basepair stability for mismatches or small insertion/deletions and large insertion/deletion loops, respectively (7,8). Part of this specialization appears to be the ability of Msh2-Msh6 to stay in tighter contact and diffuse slower on DNA as it scans for small discrepancies compared with Msh2-Msh3, which exhibits faster diffusion and more hopping (dissociation) events as it scans for large discrepancies in the double helix (71).
Due to its asymmetric ATPase activity, the MutS dimer can adopt nine different nucleotide-liganded states. Some of these states are coupled to specific MutS actions on DNA, e.g., after mismatch recognition, MutS converts to an AATP-BATP state that can interact with MutL (74,75). While the computationally intensive full 10 μs simulations were performed with MutS-T-bulge and MutS-homoduplex complexes in AADP-B0 state, we also simulated substate 6 of MutS-homoduplex for 1 μs each in the remaining eight nucleotide-liganded states and found several capable of transitioning between substates 6 and 9. It is possible that the DNA binding and ATPase activities are uncoupled in the mismatch search phase such that the properties of DNA rather than the ATPase reaction govern MutS actions as it diffuses along DNA to find an error. Once a mismatch is recognized, the two activities may be tightly coupled again to trigger ADP-ATP exchange and stabilize MutS in the AATP-BATP state. Finally, while the precise mechanism of allosteric communication between the distant mismatch binding and ATPase sites is unclear, many of the MutS residues that contact homoduplex DNA identified in this study are also part of a coevolved network of contiguous residues that connect the two sites and could modulate their activities (Asp-38, Arg-76, Arg-110, Asn-443, and Arg-475) (76). This network, identified by statistical coupling analysis, also contains residues that contact kinked DNA in the mismatch-recognition complex, including Phe-39 and Glu-41. It is an intriguing possibility that distinct but overlapping subnetworks could convey information specific to the mismatch search, recognition, and repair initiation phases of mismatch repair between the DNA binding and ATPase sites. As more dynamic views of MutS actions on DNA become available from structural and computational studies, we can look forward to higher-resolution understanding of the DNA mismatch repair mechanism.
Author contributions
A.J., K.M.T., M.M.H., and D.L.B. conceived of the research. A.J. performed all the MD simulations and data analysis. All authors helped interpret the data. A.J. and M.M.H. wrote the paper.
Acknowledgments
This work was supported by National Institutes of Health (R15 GM144870 to D.L.B., R15 GM128102 to K.M.T., and R15 GM114743 to M.M.H.). In addition, the work utilized high-performance computing resources at Wesleyan University that were supported by National Science Foundation (CNS-0619508 and CNS-0959856). We thank Prof. Michel Weir, Prof. David Langley, Anastasia Saar, and In Sub Mark Han for helpful discussions. We also thank Henk Meij for technical assistance with the high-performance computing cluster.
Declaration of interests
The authors declare no conflict of interest.
Editor: Jason Kahn.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.06.006.
Contributor Information
Abhilash Jayaraj, Email: abhilash.124@gmail.com.
Manju M. Hingorani, Email: mhingorani@wesleyan.edu.
Supporting material
References
- 1.Kunkel T.A., Erie D.A. Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 2015;49:291–313. doi: 10.1146/annurev-genet-112414-054722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiricny J. Postreplicative mismatch repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spies M., Fishel R. Mismatch repair during homologous and homeologous recombination. Cold Spring Harb. Perspect. Biol. 2015;7:a022657. doi: 10.1101/cshperspect.a022657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Z., Li X., et al. Zhang D. DNA mismatch repair and the DNA damage response. Food Chem. 2016;202:94–98. doi: 10.1016/j.dnarep.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obmolova G., Ban C., et al. Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 6.Lamers M.H., Perrakis A., et al. Sixma T.K. The crystal structure of DNA mismatch repair protein MutS binding to a G· T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 7.Warren J.J., Pohlhaus T.J., et al. Beese L.S. Structure of the human MutSα DNA lesion recognition complex. Mol. Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 8.Gupta S., Gellert M., Yang W. Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat. Struct. Mol. Biol. 2011;19:72–78. doi: 10.1038/nsmb.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hingorani M.M. Mismatch binding, ADP–ATP exchange and intramolecular signaling during mismatch repair. DNA Repair. 2016;38:24–31. doi: 10.1016/j.dnarep.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gradia S., Subramanian D., et al. Fishel R. hMSH2–hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 11.Lee J.-B., Cho W.-K., et al. Fishel R. Single-molecule views of MutS on mismatched DNA. DNA Repair. 2014;20:82–93. doi: 10.1016/j.dnarep.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma A., Doucette C., et al. Hingorani M.M. Slow conformational changes in MutS and DNA direct ordered transitions between mismatch search, recognition and signaling of DNA repair. J. Mol. Biol. 2013;425:4192–4205. doi: 10.1016/j.jmb.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borsellini A., Kunetsky V., et al. Lamers M.H. Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair. Nat. Struct. Mol. Biol. 2022;29:59–66. doi: 10.1038/s41594-021-00707-1. [DOI] [PubMed] [Google Scholar]
- 14.Kadyrov F.A., Dzantiev L., et al. Modrich P. Endonucleolytic function of MutLα in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 15.Acharya S., Foster P.L., et al. Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol. Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 16.Schofield M.J., Nayak S., et al. Hsieh P. Interaction of Escherichia coli MutS and MutL at a DNA mismatch. J. Biol. Chem. 2001;276:28291–28299. doi: 10.1074/jbc.M103148200. [DOI] [PubMed] [Google Scholar]
- 17.Grilley M., Griffith J., Modrich P. Bidirectional excision in methyl-directed mismatch repair. J. Biol. Chem. 1993;268:11830–11837. [PubMed] [Google Scholar]
- 18.Constantin N., Dzantiev L., et al. Modrich P. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeon Y., Kim D., et al. Lee J.-B. Dynamic control of strand excision during human DNA mismatch repair. Proc. Natl. Acad. Sci. USA. 2016;113:3281–3286. doi: 10.1073/pnas.1523748113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch H.T., Snyder C.L., et al. Hitchins M.P. Milestones of Lynch syndrome: 1895–2015. Nat. Rev. Cancer. 2015;15:181–194. doi: 10.1038/nrc3878. [DOI] [PubMed] [Google Scholar]
- 21.Peltomäki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J. Clin. Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 22.Natrajan G., Lamers M.H., et al. Sixma T.K. Structures of Escherichia coli DNA mismatch repair enzyme MutS in complex with different mismatches: a common recognition mode for diverse substrates. Nucleic Acids Res. 2003;31:4814–4821. doi: 10.1093/nar/gkg677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones M., Wagner R., Radman M. Repair of a mismatch is influenced by the base composition of the surrounding nucleotide sequence. Genetics. 1987;115:605–610. doi: 10.1093/genetics/115.4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazurek A., Johnson C.N., et al. Fishel R. Sequence context effect for hMSH2-hMSH6 mismatch-dependent activation. Proc. Natl. Acad. Sci. USA. 2009;106:4177–4182. doi: 10.1073/pnas.0808572106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorman J., Chowdhury A., et al. Greene E.C. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol. Cell. 2007;28:359–370. doi: 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu R., DeRocco V.C., et al. Weninger K.R. Large conformational changes in MutS during DNA scanning, mismatch recognition and repair signalling. EMBO J. 2012;31:2528–2540. doi: 10.1038/emboj.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W. Structure and mechanism for DNA lesion recognition. Cell Res. 2008;18:184–197. doi: 10.1038/cr.2007.116. [DOI] [PubMed] [Google Scholar]
- 28.Isaacs R.J., Spielmann H.P. A model for initial DNA lesion recognition by NER and MMR based on local conformational flexibility. DNA Repair. 2004;3:455–464. doi: 10.1016/j.dnarep.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Bouchal T., Durník I., Kulhánek P. Bending of Canonical and G/T Mismatched DNAs. J. Chem. Inf. Model. 2021;61:6000–6011. doi: 10.1021/acs.jcim.1c00731. [DOI] [PubMed] [Google Scholar]
- 30.Fernandez-Leiro R., Bhairosing-Kok D., et al. Lamers M.H. The selection process of licensing a DNA mismatch for repair. Nat. Struct. Mol. Biol. 2021;28:373–381. doi: 10.1038/s41594-021-00577-7. [DOI] [PubMed] [Google Scholar]
- 31.Bhairosing-Kok D., Groothuizen F.S., et al. Sixma T.K. Sharp kinking of a coiled-coil in MutS allows DNA binding and release. Nucleic Acids Res. 2019;47:8888–8898. doi: 10.1093/nar/gkz649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bank P.D. Protein data bank. Nature New Biol. 1971;233:223. [Google Scholar]
- 34.Alani E., Lee J.Y., et al. Yang W. Crystal structure and biochemical analysis of the MutS· ADP· beryllium fluoride complex suggests a conserved mechanism for ATP interactions in mismatch repair. J. Biol. Chem. 2003;278:16088–16094. doi: 10.1074/jbc.M213193200. [DOI] [PubMed] [Google Scholar]
- 35.Lebbink J.H.G., Fish A., et al. Sixma T.K. Magnesium coordination controls the molecular switch function of DNA mismatch repair protein MutS. J. Biol. Chem. 2010;285:13131–13141. doi: 10.1074/jbc.M109.066001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 2016;86:5–6. doi: 10.1002/cpps.20. [DOI] [PubMed] [Google Scholar]
- 37.Junop M.S., Obmolova G., et al. Yang W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell. 2001;7:1–12. doi: 10.1016/s1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- 38.Li P., Merz K.M., Jr. 2016. MCPB. Py: A python Based Metal Center Parameter Builder. [DOI] [PubMed] [Google Scholar]
- 39.Case D.A., Ben-Shalom I.Y., et al. Goetz A.W. University of California; San Francisco: 2018. AMBER 2018. [Google Scholar]
- 40.Frisch, M.J., G.W. Trucks, … Hratch. Gaussian 16, Revision C.01. .
- 41.Anandakrishnan R., Aguilar B., Onufriev A.V. H++ 3.0: automating p K prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 2012;40:W537–W541. doi: 10.1093/nar/gks375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maier J.A., Martinez C., et al. Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galindo-Murillo R., Robertson J.C., et al. Cheatham T.E., III Assessing the current state of amber force field modifications for DNA. J. Chem. Theory Comput. 2016;12:4114–4127. doi: 10.1021/acs.jctc.6b00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jorgensen W.L., Chandrasekhar J., et al. Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 45.Essmann U., Perera L., et al. Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 46.Darden T., York D., Pedersen L. Particle mesh Ewald: An N· log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 47.Crowley M., Darden T., et al. Deerfield Ii D. Adventures in improving the scaling and accuracy of a parallel molecular dynamics program. J. Supercomput. 1997;11:255–278. [Google Scholar]
- 48.Ryckaert J.-P., Ciccotti G., Berendsen H.J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 49.Berendsen H.J.C., Postma J.P.M., et al. Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 50.Salomon-Ferrer R., Götz A.W., et al. Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 51.Götz A.W., Williamson M.J., et al. Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J. Chem. Theory Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 53.Thayer K.M., Lakhani B., Beveridge D.L. Molecular dynamics–Markov state model of protein ligand binding and Allostery in CRIB-PDZ: conformational selection and induced fit. J. Phys. Chem. B. 2017;121:5509–5514. doi: 10.1021/acs.jpcb.7b02083. [DOI] [PubMed] [Google Scholar]
- 54.Blanchet C., Pasi M., et al. Lavery R. CURVES+ web server for analyzing and visualizing the helical, backbone and groove parameters of nucleic acid structures. Nucleic Acids Res. 2011;39:W68–W73. doi: 10.1093/nar/gkr316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Janert P.K. Simon and Schuster; 2016. Gnuplot in Action: Understanding Data with Graphs. [Google Scholar]
- 56.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 57.DeLano W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsletter on protein crystallography. 2002;40:82–92. [Google Scholar]
- 58.Pettersen E.F., Goddard T.D., et al. Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 59.Antony E., Khubchandani S., et al. Hingorani M.M. Contribution of Msh2 and Msh6 subunits to the asymmetric ATPase and DNA mismatch binding activities of Saccharomyces cerevisiae Msh2–Msh6 mismatch repair protein. DNA Repair. 2006;5:153–162. doi: 10.1016/j.dnarep.2005.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Antony E., Hingorani M.M. Asymmetric ATP binding and hydrolysis activity of the Thermus aquaticus MutS dimer is key to modulation of its interactions with mismatched DNA. Biochemistry. 2004;43:13115–13128. doi: 10.1021/bi049010t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Antony E., Hingorani M.M. Mismatch recognition-coupled stabilization of Msh2-Msh6 in an ATP-bound state at the initiation of DNA repair. Biochemistry. 2003;42:7682–7693. doi: 10.1021/bi034602h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mukherjee S., Law S.M., Feig M. Deciphering the mismatch recognition cycle in MutS and MSH2-MSH6 using normal-mode analysis. Biophys. J. 2009;96:1707–1720. doi: 10.1016/j.bpj.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mukherjee S., Feig M. Conformational change in MSH2-MSH6 upon binding DNA coupled to ATPase activity. Biophys. J. 2009;96:L63–L65. doi: 10.1016/j.bpj.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalnik M.W., Norman D.G., et al. Patel D.J. Conformational transitions in thymidine bulge-containing deoxytridecanucleotide duplexes. Role of flanking sequence and temperature in modulating the equilibrium between looped out and stacked thymidine bulge states. J. Biol. Chem. 1990;265:636–647. [PubMed] [Google Scholar]
- 65.Yang Y., Sass L.E., et al. Erie D.A. Determination of protein–DNA binding constants and specificities from statistical analyses of single molecules: MutS–DNA interactions. Nucleic Acids Res. 2005;33:4322–4334. doi: 10.1093/nar/gki708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rossetti G., Dans P.D., et al. Orozco M. The structural impact of DNA mismatches. Nucleic Acids Res. 2015;43:4309–4321. doi: 10.1093/nar/gkv254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharma M., Predeus A.V., et al. Feig M. DNA bending propensity in the presence of base mismatches: implications for DNA repair. J. Phys. Chem. B. 2013;117:6194–6205. doi: 10.1021/jp403127a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang H., Yang Y., et al. Erie D.A. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc. Natl. Acad. Sci. USA. 2003;100:14822–14827. doi: 10.1073/pnas.2433654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tessmer I., Yang Y., et al. Erie D.A. Mechanism of MutS searching for DNA mismatches and signaling repair. J. Biol. Chem. 2008;283:36646–36654. doi: 10.1074/jbc.M805712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolf A., Kirschner K.N. Principal component and clustering analysis on molecular dynamics data of the ribosomal L11· 23S subdomain. J. Mol. Model. 2013;19:539–549. doi: 10.1007/s00894-012-1563-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pal A., Greenblatt H.M., Levy Y. Prerecognition diffusion mechanism of human DNA mismatch repair proteins along DNA: Msh2-Msh3 versus Msh2-Msh6. Biochemistry. 2020;59:4822–4832. doi: 10.1021/acs.biochem.0c00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Plazzer J.-P., Sijmons R.H., et al. Macrae F. The InSiGHT database: utilizing 100 years of insights into Lynch syndrome. Fam. Cancer. 2013;12:175–180. doi: 10.1007/s10689-013-9616-0. [DOI] [PubMed] [Google Scholar]
- 73.Bouchal T., Durník I., et al. Kulhánek P. Importance of base-pair opening for mismatch recognition. Nucleic Acids Res. 2020;48:11322–11334. doi: 10.1093/nar/gkaa896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mazur D.J., Mendillo M.L., Kolodner R.D. Inhibition of Msh6 ATPase activity by mispaired DNA induces a Msh2 (ATP)-Msh6 (ATP) state capable of hydrolysis-independent movement along DNA. Mol. Cell. 2006;22:39–49. doi: 10.1016/j.molcel.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 75.Hao P., LeBlanc S.J., et al. Weninger K.R. Recurrent mismatch binding by MutS mobile clamps on DNA localizes repair complexes nearby. Proc. Natl. Acad. Sci. USA. 2020;117:17775–17784. doi: 10.1073/pnas.1918517117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lakhani B., Thayer K.M., et al. Beveridge D.L. Evolutionary covariance combined with molecular dynamics predicts a framework for allostery in the MutS DNA mismatch repair protein. J. Phys. Chem. B. 2017;121:2049–2061. doi: 10.1021/acs.jpcb.6b11976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The observed motions are primarily separation of MutS clamp domains (IV) and lateral movement of DNA.
The observed motions are primarily unbending of homoduplex DNA from kinked to straight conformation.
Key contact residues are shown (described in Fig. 7). The overall DNA shape, bendability, and local base stacking and pairing stability could be assessed through dynamic, concerted interactions of MutS with the sugar-phosphate backbone and bases over one to two helical turns.