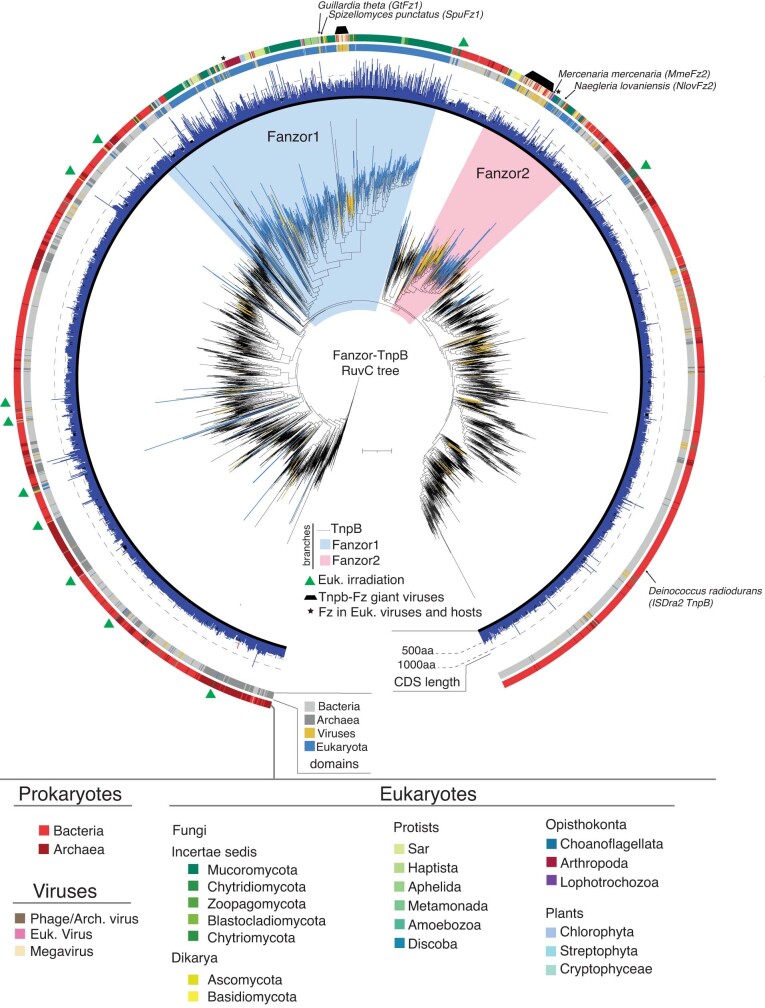
Extended Data Fig. 1. Phylogenetic tree of Fanzor and TnpB.
Phylogenetic tree built from the RuvC region of hits detected from structural and profile mining of Fanzor. Blue, black and yellow leaves indicate the domain annotation of the contig where the hit is found respectively eukaryotes, viruses and prokaryotes. Fanzor1 and Fanzor2 clades are shown respectively in blue and pink. Fanzors and TnpB of interest are indicated by arrows. The bars forming the blue inner ring are proportional to the size of the Fanzors in aa as annotated in the database. The middle ring indicates the domains of life from which the Fanzor/TnpB is found (light gray: bacteria, dark gray: archaea, yellow: viruses, blue: eukaryotes). The outer ring displays the taxonomy of the organism in which the Fanzor/TnpB is found (red: bacteria, dark red: archaea, brown: phage and archaeal viruses, pink: eukaryotic viruses, beige: giant viruses, dark green to yellow gradient: fungi, light green gradient: protists, dark blue: opisthokonta (choanoflagellata), crimson: arthropoda, purple: mollusks, and light blue to dark blue gradient: plants with Chlorophyta, Streptophyta, and Cryptophyceae. The green triangles in the outer ring indicate clusters of hits from contigs annotated to be eukaryotes and represent putative eukaryotic radiations. Black trapezoid shapes indicate the two branches containing giant viruses and bacterial hosts.