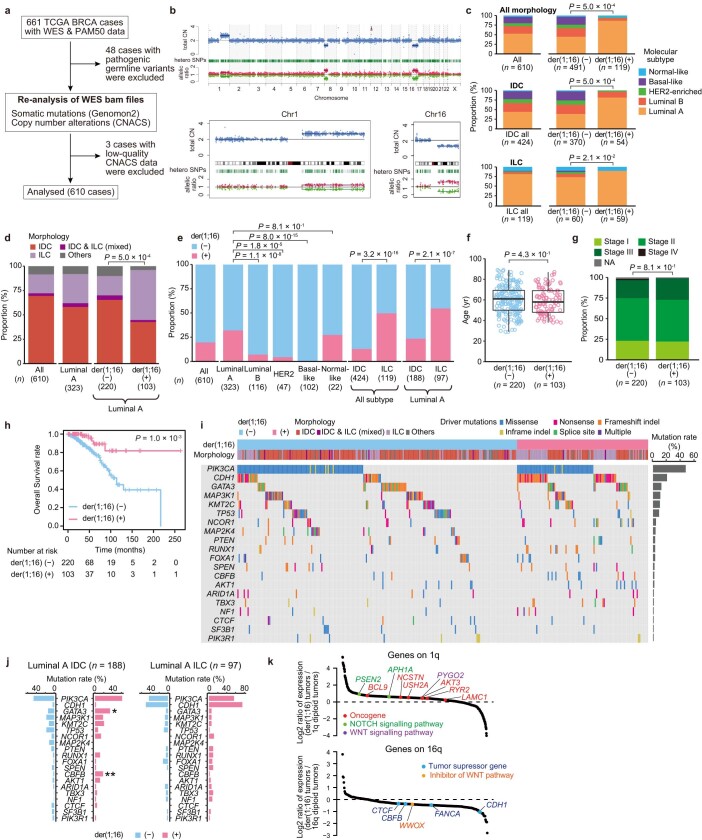
Extended Data Fig. 10. Features of der(1;16)(+) breast cancer in TCGA cohort.
a, Schema of re-analysis of TCGA breast cancer (BRCA) cohort (Methods). b, Representative copy number plots in der(1;16)(+) cancers analysed using CNACS. c, Distribution of PAM50 molecular subtypes in all cancers (n = 610), IDC (n = 424), and ILC (n = 119), with and without der(1;16), with P-values from two-sided Fisher’s exact test. d, Distribution of morphology, which was compared between der(1;16)(−) and der(1;16)(+) Luminal A cancers using two-sided Fisher’s exact test. e, Proportion of der(1;16)(+) cancer in each PAM50 molecular subtype, and IDC and ILC in all subtypes and Luminal A, respectively, with P-values from two-sided Fisher’s exact test. f,g, Distribution of age (f) and stage (g) in der(1;16)(−) and der(1;16)(+) Luminal A cancer cases, with P-values from two-sided Mann–Whitney U test and Fisher’s exact test, respectively. Box plots show the median, first and third quartiles, with whiskers that extend to the furthest value within a 1.5× interquartile range. h, Kaplan-Meier survival analysis of der(1;16)(−) and der(1;16)(+) Luminal A cancer cases with P-values from two-sided log-rank test. i, Landscape of driver mutations in der(1;16)(−) and der(1;16)(+) Luminal A cancers (n = 220 and 103, respectively). Frequency of each mutation is indicated on the right-hand side. j, Frequency of driver mutations among der(1;16)(−) and der(1;16)(+) cancers in Luminal A IDC (n = 188) and Luminal A ILC (n = 97), respectively. **, q<0.05; *, q<0.1 (from two-sided Fisher’s exact test with Benjamini–Hochberg adjustment). k, Ratio of average expression of genes on 1q in der(1;16)(+) Luminal A tumours (n = 103) to those in 1q-diploid Luminal A tumours (n = 77) was depicted on the top, and the ratio of genes on 16q in der(1;16)(+) Luminal A tumours (n = 51) to those in 16q-diploid Luminal A tumours (n = 83) was depicted on the bottom. Coloured dots indicate known oncogenes, tumour suppressor genes, or other genes on the NOTCH and WNT signalling pathways.