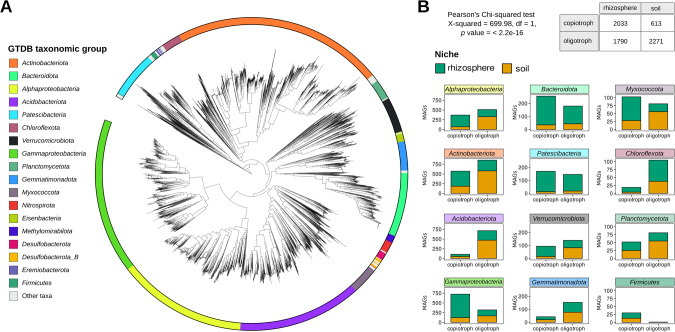
Fig. 2. MAGs taxonomy, niche, and growth rate status.
A Unrooted maximum-likelihood phylogenetic tree inferred from multiple sequence alignments of GTDB bacterial marker genes from MAGs. The tree was generated with GTDB-Tk and displayed using iTol [80]. B MAGs are classified according to their isolation biome and growth rate status (copiotroph or oligotroph) for each of the main GTDB taxa. MAGs included members affiliated with the Actinobacteriota (1453), Proteobacteria (including 1063 MAGs from Gammaproteobacteria and 869 MAGs from Alphaproteobacteria), Acidobacteriota (850), Bacteroidetes (440), Patescibacteria (322), Verrucomicrobiota (248), Gemmatimonadota (200), Myxococcota (187), Planctomycetota (133), Chloroflexota (127), Nitrospirota (76), Eisenbacteria (56), Methylomirabilota (51), Desulfobacterota (47), Desulfobacterota_B (45), and Firmicutes (36), among others (Supplementary Table 4).