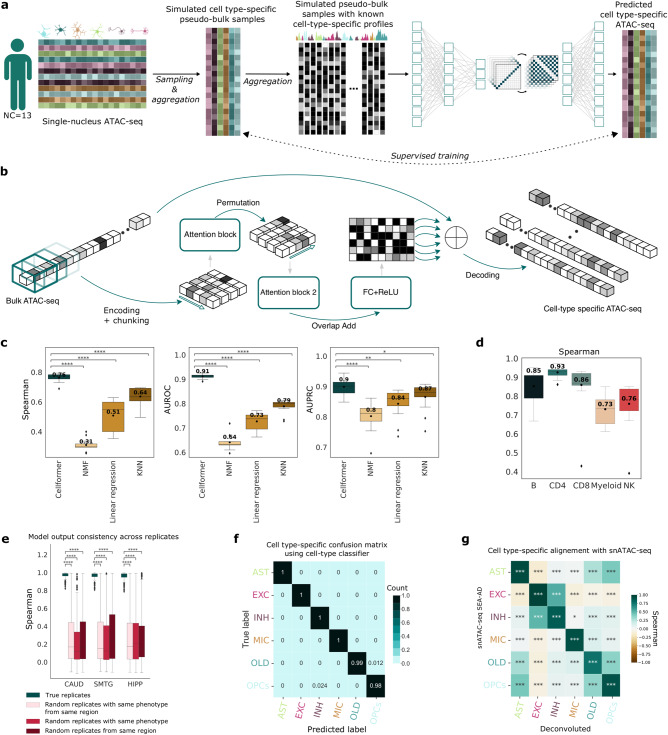
Fig. 2. Cellformer: model training, design, and evaluation.
a A synthetic dataset of simulated bulk samples was generated from previously published single-cell ATAC-seq from 13 normal controls7. Cell type-specific pseudo-bulk samples were generated by aggregating snATAC-seq data, revealing the ground truth cell type-specific composition. Simulated cell-specific pseudo-bulk samples were further aggregated to generate pseudo-bulk samples, which are Cellformer’s input. This dataset was used to train Cellformer to minimize the reconstruction error between predicted and ground truth cell type-specific ATAC-seq (Created with Biorender.com). b Cellformer leverages a dual-path strategy to process both intra and inter-chromosome interaction, enabling full genome deconvolution. P values were derived using a two-sided Wilcoxon’s test after multi-testing correction. c Cellformer was evaluated using the leave-one-subject-out strategy. It outperformed other multi-output regression models, notably linear regression, KNN and an unsupervised approach (NMF) used previously to estimate cellular composition across the (n = 6) different cell types. P values were derived using a two-sided Wilcoxon’s test after multi-testing correction. d Cellformer successfully deconvoluted leave-one-out cross-validated PBMC in-silico bulk ATAC-seq data from different datasets (n = 18 samples), predicting cell type-specific expression of five main cell types (B cell, T cell-CD4+ (CD4), T cell-CD8+ (CD8), Myeloid and NK cells). P values were derived using a two-sided Wilcoxon’s test after multi-testing correction. e Quality of the Cellformer’s predictions was assessed by comparing technical replicate cell type-specific expression (n = 36 samples, see Fig. 1). Cellformer generated outputs that are highly consistent between true technical replicates, exhibiting a correlation coefficient (>0.9) significantly higher than with random replicates. (Two-sided Wilcoxon’s test after multi-testing correction) f Cellformer output preserves cell type signature across 6 cell types: astrocytes (AST), microglia (MIC), oligodendrocytes (OLD), and oligodendrocyte progenitor cells (OPCs), and 2 major classes of neurons, excitatory (EXC) and inhibitory (INH). An external cell classifier trained on single-cell data from NC samples was used to assess the cell type-specific ATAC-seq quality. The confusion matrix computed between the cell classifier and Cellformer predictions showed almost perfect agreement, highlighting its capacity to preserve the cell type signature. g Cellformer validation was performed by comparing RAD cell type-specific expression from SMTG with RAD single-cell ATAC-seq expression from SEA-AD using a two-sided Spearman correlation. Significant high correlations were obtained within the same cell type between the two datasets. Spearman correlation coefficient order between cell types was consistent with biological knowledge: a high correlation was found between neuron types and between OLD and OPCs. All box plots show the median (middle line), interquartile range (bottom and upper edges), and the minimum and maximum values of the distribution (whiskers). *P value < 0.05, **P value < 0.01, ***P value < 0.001, ****P value < 0.0001.