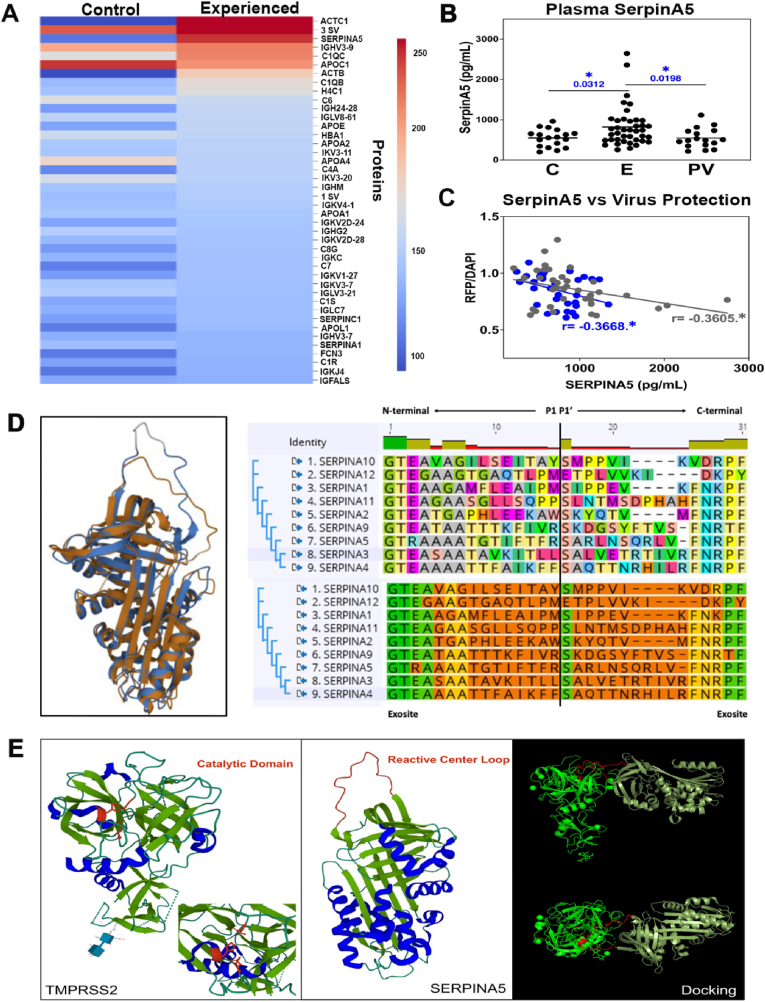
Fig. 3.
Experienced meditators have elevated levels of plasma SERPINA5. a, Pseudovirus interactome co-immunoprecipitated from control and experienced post-meditation plasma using spike antibody. b, SERPINA5 concentration (pg/mL) in control and experienced post-meditation plasma (control n=18, experienced n=40). c, Correlation analysis between plasma SERPINA5 concentration (pg/mL) and viral infection protection (RFP/DAPI) in novice (n = 37, gray dots) and experienced (n = 33, blue dots) meditators. r, Pearson's correlation value, (*p < 0.05). d, Ribbon diagram of SERPINA1 (3CWM) and SERPINA5 (2OL2) superimposed on one another. SERPINA1 and SERPINA5 were aligned with the RCSB pairwise structure alignment tool (Berman et al., 2000) using the iFACTCAT(rigid) algorithm (Li et al., 2020). The reactive center loop (RCL) is depicted in the right panel for all inhibitory clade A SERPINs. The top alignment represents color by amino acid while the bottom alignment represents similarity as calculated by Blosum62 score matrix in Geneious (G. 11.1.5). Green amino acids represent 80–100% similarity, yellow amino acids represent 60–80% similarity, and orange amino acids represent less than 60% similarity. Both N- and C-terminal exocites are intact as well as a central Ser (P1|P1’) with the exception of SERPINA12. Alignment scheme adapted from Sanrattana et al. (Sanrattana et al., 2019). e, Ribbon diagrams of TMPRSS2 (PDB 7MEQ) (catalytic domain, depicted in red; left panel), SERPINA5 (2OL2) with RCL (depicted in red; middle panel). Beta sheets are shown in green, α-helices in blue. Potential TMPRSS2/SERPINA5 docking structures are indicated (right panel). Firedock global energy (3.21), attractive VdW (−1.35), Repulsive VdW (0.49), ACE (−0.46), HB (0.0). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)