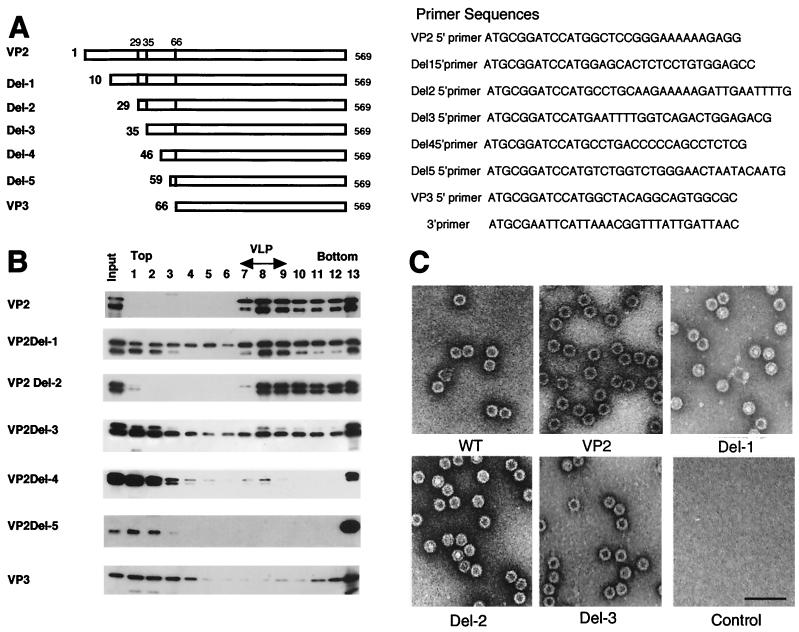
FIG. 1.
Analysis of VLP formation by N-terminal deletion analysis of VP2. (A) Schematic representation of truncated VP2 proteins. The numbers on the left indicate the first amino acid of each VP2 protein. (B) Sucrose density gradient analysis of N-terminal deletion mutants of VP2. A series of N-terminal deletion mutants were expressed by using recombinant baculoviruses. VLP formation was analyzed by using sucrose density gradient ultracentrifugation. Each fraction was subjected to SDS-PAGE followed by immunoblotting with anti-VP3 serum. Smaller proteins detected in each sample were degradation products of VP2 as determined by amino acid sequence analyses of the smaller proteins. Fraction numbers (from the top to the bottom of the gradient) are shown at the top of the figure. VLP-containing fractions were identified by electron microscopy. (C) Electron microscopy. Purified wild-type empty capsid and VLPs from recombinant baculovirus-infected cells were negatively stained and observed by electron microscopy. Wild-type empty capsid was prepared as described by Ruffing et al. (12). Bar, 100 nm.