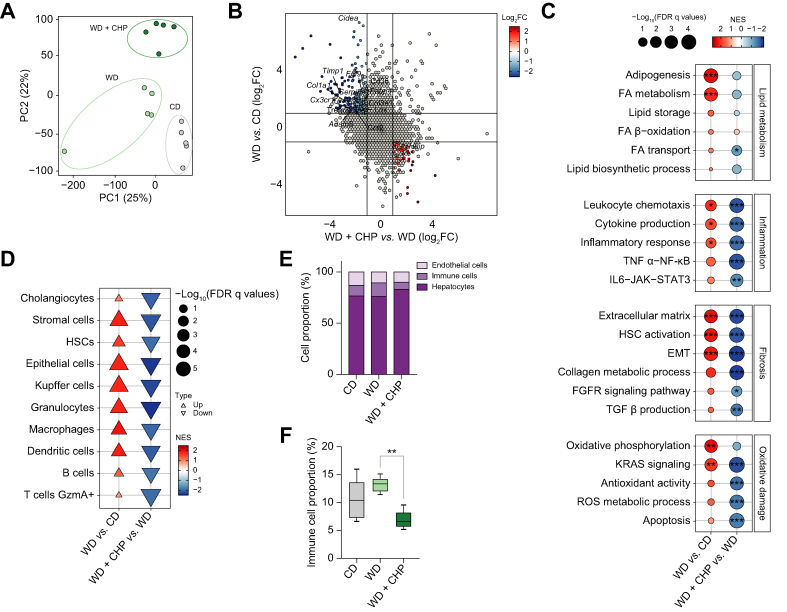
Fig. 3.
Transcriptomic signature of CHP in liver.
(A) PCA plot of the RNA-seq profiles of the 3 groups. (B) Scatter plot shows the effects of WD compared to CD and the therapeutic effects of CHP on gene expression. The significant genes that show a reversal of the effect of WD compared with CHP treatment are highlighted in blue (upregulated upon WD) and red (upregulated upon treatment). Representative genes are annotated. (C) GSEA of disease (WD) and treatment (CHP) effects on gene expression. Gene sets are grouped into 4 categories: lipid metabolism, inflammation, fibrosis, and oxidative damage. ∗Q <0.05; ∗∗Q <0.01; ∗∗∗Q <0.001. (D) Cell type enrichment analysis of disease (WD) and treatment (CHP) effects on gene expression. (E) Single cell deconvolution results obtained from integrating the FACS scRNA data available under the GSE109774 accession and our bulk RNA-seq dataset, showing the relative depletion of immune cells upon CHP treatment. (F) Single-cell deconvolution estimated grouped cell proportions of immune cells in different conditions. Whiskers in boxplots represent the minimum to maximum range. One-way ANOVA, followed by Dunnett’s multiple comparison test vs. WD group was used for statistical analysis. ∗∗p <0.01. ANOVA, analysis of variance; CD, chow diet; CHP; cyclo(His-Pro); EMT, epithelial–mesenchymal transition; FA, fatty acids; FC, fold change; FDR, false discovery rate; FGFR, fibroblast growth factor receptor; GSEA, gene set enrichment analysis; HSC, hepatic stellate cells; NES, normalized enrichment score; NF-κB, nuclear factor kappa B; PCA, principal component analysis; ROS, reactive oxygen species; TGF-β, transforming growth factor beta; TNF-α, tumour necrosis factor alpha; WD, western diet.