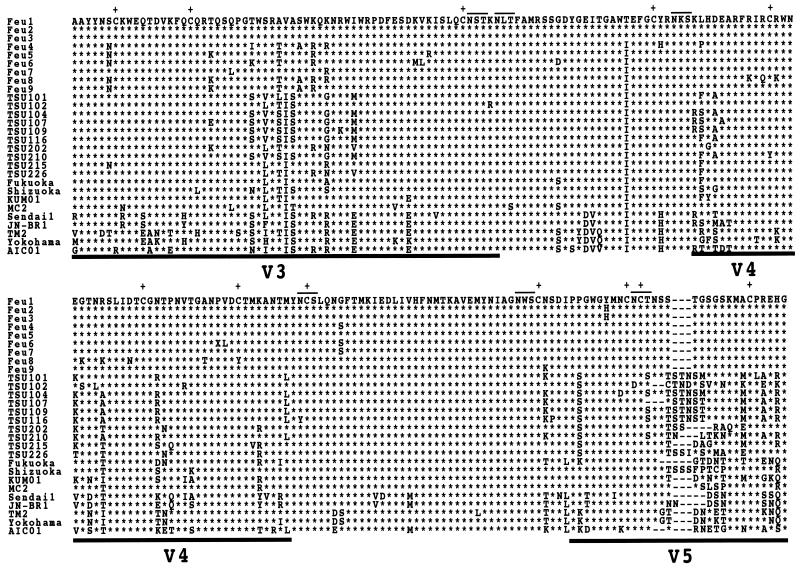
FIG. 1.
Alignment of the predicted amino acid sequences of the FIV env gene of 9 clones from a Tsushima cat (Feu1 to -9), 10 clones from domestic cats on Tsushima Island (TSU101, TSU102, TSU104, TSU107, TSU109, TSU116, TSU202, TSU210, TSU215, and TSU226), and 9 known FIV clones, by using the CLUSTAL W program. The FIV clones previously reported include Fukuoka (GenBank/EMBL/DDBJ accession no. D37815), Shizuoka (D37811), KUM01 (AB010405), MC2 (D67062), Sendai1 (D37813), JN-BR1 (D67052), TM2 (M59418), Yokohama (D37812), and AIC01 (AB010395). Asterisks denote amino acid identity with the Feu1 strain. The horizontal lines above the sequence indicate the locations of N-linked glycosylation sites. The pluses indicate the positions of cysteine residues. The dashes represent gaps introduced for optimal alignment. The thick bars under the alignment indicate the positions of the variable regions V3 to V5 as defined by Pancino et al. (21).