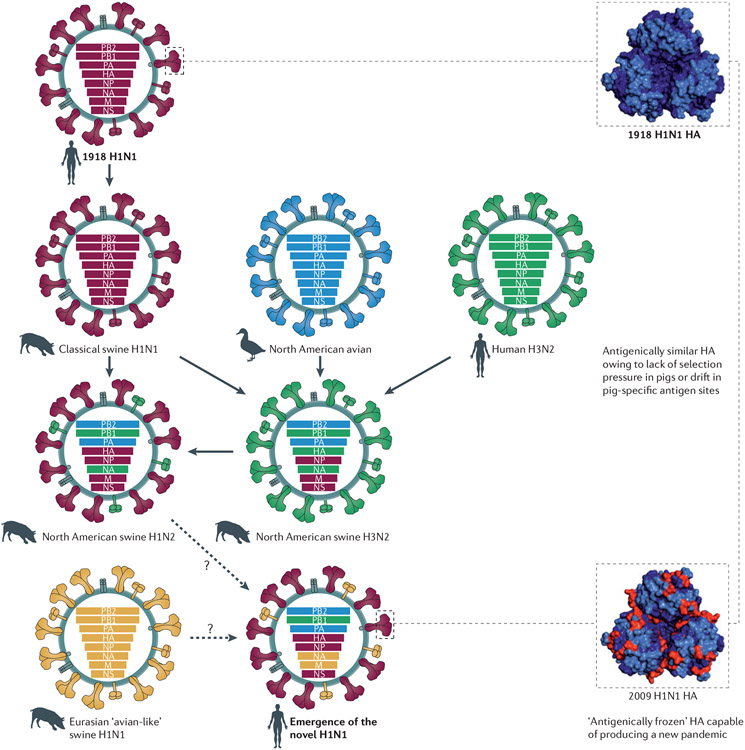
Figure 2 ∣. Emergence of an ‘antigenically frozen’ 2009 pandemic H1N1 virus.
Influenza viruses similar to the 1918 pandemic H1N1 virus became established in domestic pigs between 1918 and 1920; this lineage is referred to as the classical swine lineage. In 1979, a distinct Eurasian ‘avian-like’ H1N1 virus emerged in European pigs and has since co-circulated with the classical swine H1N1 viruses. Triple-reassortant swine origin influenza virus (SOIV) H1 viruses of different strains and subtypes (for example, H3N2 and H1N2) emerged and became predominant among North American pig herds in the 1990s. All of these viruses provided the genetic pool for the genesis of the 2009 pandemic H1N1 SOIV, possibly owing to further reassortment in pigs. Thus, the 2009 pandemic H1N1 virus is composed of PB2 and PA segments from North American avian viruses, the PB1 segment of the human H3N2 viruses, haemagglutinin (HA; of the H1 subtype), nucleoprotein (NP) and NS segments derived from classical swine H1N1 viruses, and the neuraminidase (NA; of the N1 subtype) and M segments of Eurasian ‘avian-like’ swine viruses. Sequence and antigenic analyses of the 2009 pandemic H1N1 virus show that there are similarities between the HA of this virus and that of the 1918 and human H1N1 viruses that circulated sometime between 1918 and the 1950s. The antigenic similarities between the 1918 and 2009 pandemic H1N1 viruses are represented in the crystal structure models of the trimeric configuration of the HA protein globular head, as seen from a top view. The antigenic sites of the HA proteins are shown in light blue, non-antigenic sites are shown in dark blue. The sites that differ between the 1918 and 2009 HA proteins are depicted in red.