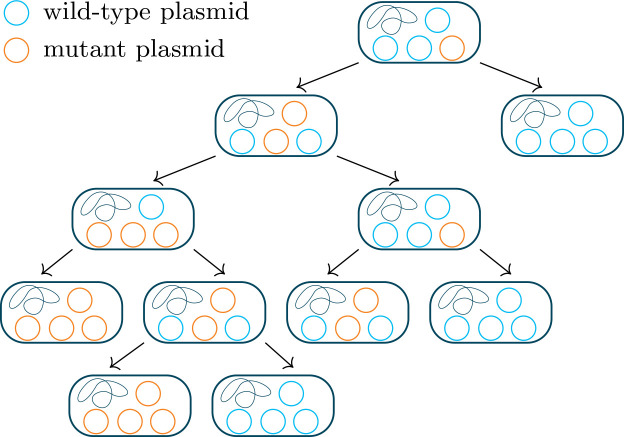
Fig. 4.
Schema of the effect of segregational drift on a novel allele located on a plasmid. A novel allele originally arises on one copy of the plasmid, and by random segregation wild-type and mutant plasmids eventually end up in different, homozygous cells. We here describe one way of modelling the process mathematically [228]. In the simplest case, cell dynamics are modelled by a birth-death process with per capita birth and death rates and , which may depend on the number of mutant plasmids in a given cell. Prior to cell division, plasmids are replicated such that the cell contains twice the original number. Two possible models for plasmid replication are ‘regular replication’, where each copy is replicated exactly once, and ‘random replication’, where copies are randomly picked for replication one by one (cf. [198]). Each daughter cell then receives half of those plasmids with segregation being random with respect to mutant and wild-type variants. With n total plasmid copies and x mutant copies after plasmid replication, the probability that one daughter cell receives and the other one mutant copies is then given by , where denotes Kronecker’s delta. Adapted from [90, 245].