Fig. 3.
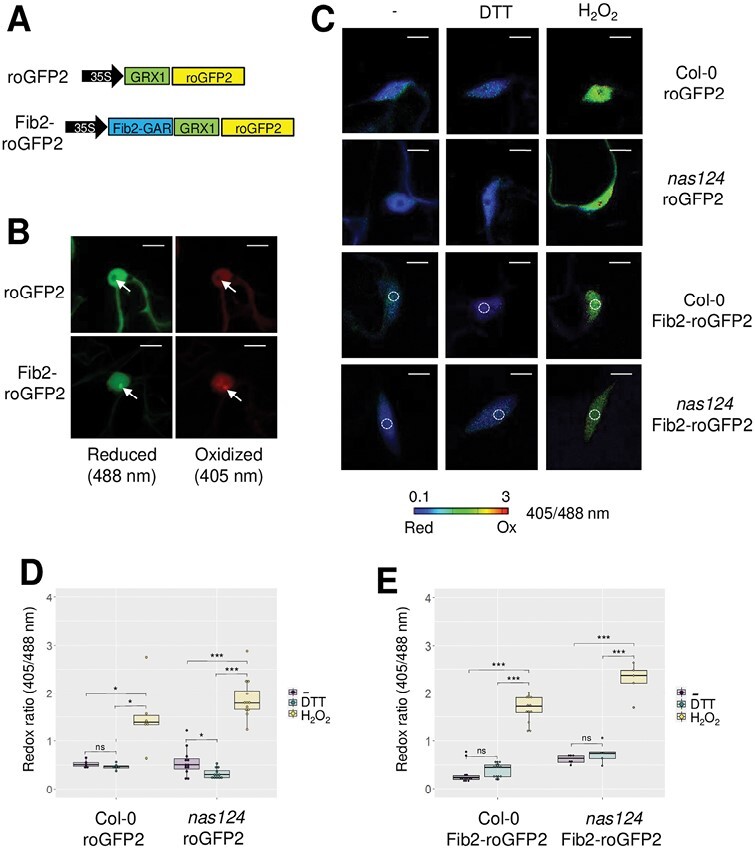
In vivo monitoring of the nucleolar/nuclear redox state in nas124. (A) Gene structure of the roGFP2 constructs used to transform WT and nas124 plants. Both constructs were under the control of the 35S-CaMV promoter. In Fib2-roGFP2, GRX1 was fused to roGFP2 as in roGFP2, but the GAR domain (1–73 N-terminal amino acids) of Fib2 was additionally fused to GRX1-roGFP2. (B) Confocal images of nuclei of WT cotyledons expressing roGFP2 or Fib2-roGFP2. RoGFP2 was excited at 488 nm or 405 nm to monitor the reduced or oxidized forms, respectively. Arrows indicate the nucleolus. Scale bars=10 µm. (C) Steady state ratio images of WT or nas124 plants expressing roGFP2 or Fib2-roGFP2 and calculated as the 405/488 nm fluorescence. To fully reduce or oxidize the sensor, seedlings were immersed in 10 mM DTT or 100 mM H2O2, respectively. Control samples were immersed in MS/2 liquid medium only. False colours indicate the fluorescence ratio on a scale from blue (reduced) to red (oxidized). Scale bars=10 µm. White circles indicate the location of the nucleolus. (D-E) Fluorescence ratio of roGFP2 (D) and Fib2-roGFP2 (E) calculated from ratio images. Ratios were calculated from six to 16 nuclei per sample. Asterisks indicate a significant difference calculated by Student’s t-test (* P≤0.05, ** P≤0.01, and *** P≤0.001, ns P>0.05).
