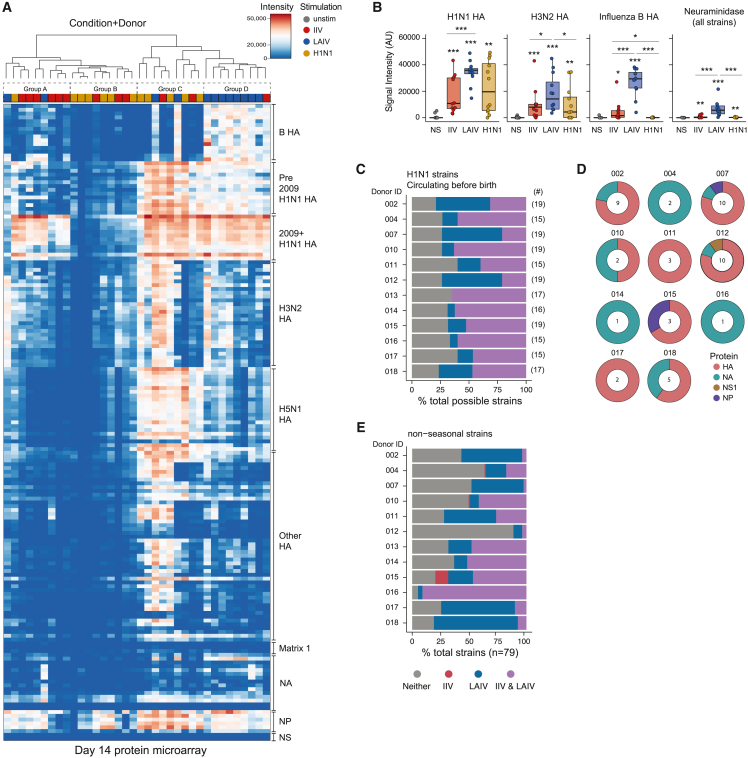
Figure 2.
Influenza vaccine modality influences antibody magnitude, specificity, and breadth
(A) Heatmap of antibody-binding magnitude against influenza proteins on a high-throughput protein microarray. Culture supernatants are from day 14 organoids. Column dendrogram represents unbiased sample grouping based on similarity; top bar color represents the antigen stimulation. Rows represent individual proteins on the microarray and were manually arranged based on influenza strain origin and protein type.
(B) Summary antibody data from the protein microarray by protein type and virus source. Data represent median values. Each point is an individual donor. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 using a Kruskal-Wallis test followed by paired Mann-Whitney U tests to compare groups. p values shown are for comparisons against the unstimulated control unless otherwise indicated by lines. Boxplots show the median, with hinges indicating the first and third quartiles and whiskers indicating the highest and lowest value within 1.5 times the interquartile range of the hinges.
(C) Detection of cross-reactive Ab production from IIV- vs. LAIV-stimulated tonsil organoids. Each donor is a row. The numbers of protein antigens from H1N1 strains that circulated prior to each donor’s birth are shown to the right of the plot. Abs produced from the organoids were classified as either present or absent in the culture supernatants.
(D) Protein targets from Ab responses unique to LAIV-stimulated organoids. NA, neuraminidase; NS, nonstructural; NP, nucleoprotein. The number of strains uniquely targeted by Abs from LAIV stimulation are shown in doughnut centers.
(E) Organoid Ab responses to nonseasonal influenza strain proteins on the microarray. Ab presence or absence was classified and plotted as in (C). n = 12 donors for all analyses (1 experiment). See also Tables 1, S1, and S6.