Figure 3.
Influenza vaccine modalities elicit distinct antigen-specific B cell populations
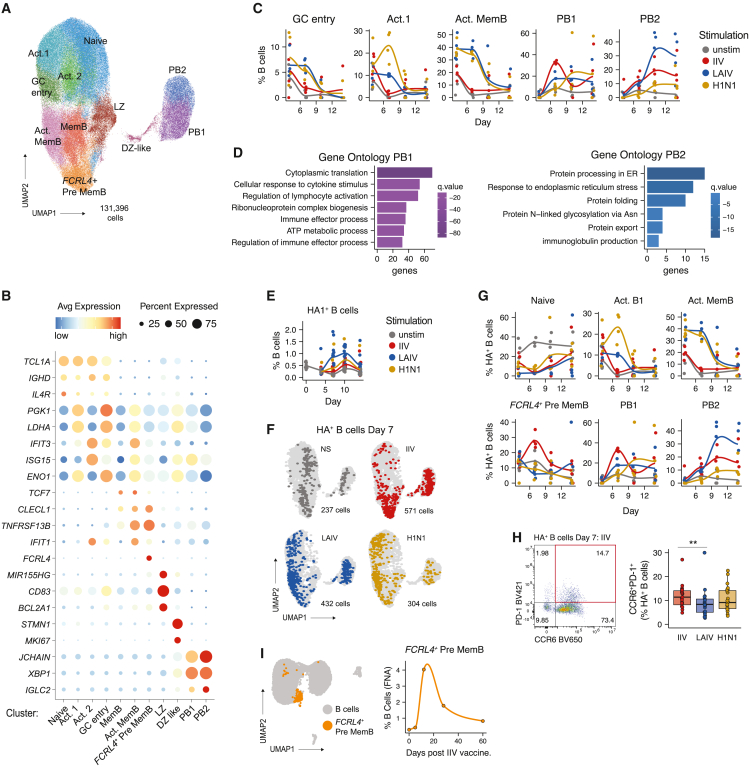
(A) Combined uniform manifold approximation and projection (UMAP) of scRNA-seq data from immune organoid B cells. Cells from days 4, 7, 10, and 14 and all stimulation conditions are shown in aggregate. Clusters are represented by different colors and labels were manually annotated based on transcriptional and protein profiles. n = 4 donors (1 experiment).
(B) Bubble plot of select marker genes for each B cell cluster. Bubble size indicates the frequency of cells with gene expression and color represents level of RNA expression.
(C) B cell scRNA-seq cluster frequencies over time for each stimulation. Each donor is a point. n = 4 donors (1 experiment).
(D) Gene ontology analysis comparing PB1 and PB2 transcriptional profiles.
(E) Kinetics of HA+ B cell frequencies as determined by flow cytometry. n = 12 donors (1 experiment).
(F) Distribution of day 7 HA+ B cells (colored points) on the total B cell (light gray) UMAP. n = 4 donors (1 experiment).
(G) Transcriptional profiles (cluster identity) and kinetics of HA+ B cells. Frequencies shown were calculated out of total HA+ B cells. n = 4 donors (1 experiment).
(H) Representative flow cytometry staining (left) and quantification (right) of CCR6+PD-1+ HA+ pre-memory B cells on day 7 poststimulation. n = 23 donors (3 experiments combined). Paired Mann-Whitney U tests were used to compare groups; ∗∗p < 0.01
(I) Analysis of public data from Turner et al.20 UMAP (left) and quantification (right) of FCRL4+ memory B cells in fine needle aspirates (FNAs) of an adult vaccinated with IIV. Data were derived from a public scRNA-seq dataset (GSE148633). n = 1 donor analyzed before vaccination and on days 5, 12, 28, and 60 postimmunization. See also Tables 1 and S2 and Figure S3.