FIG 7.
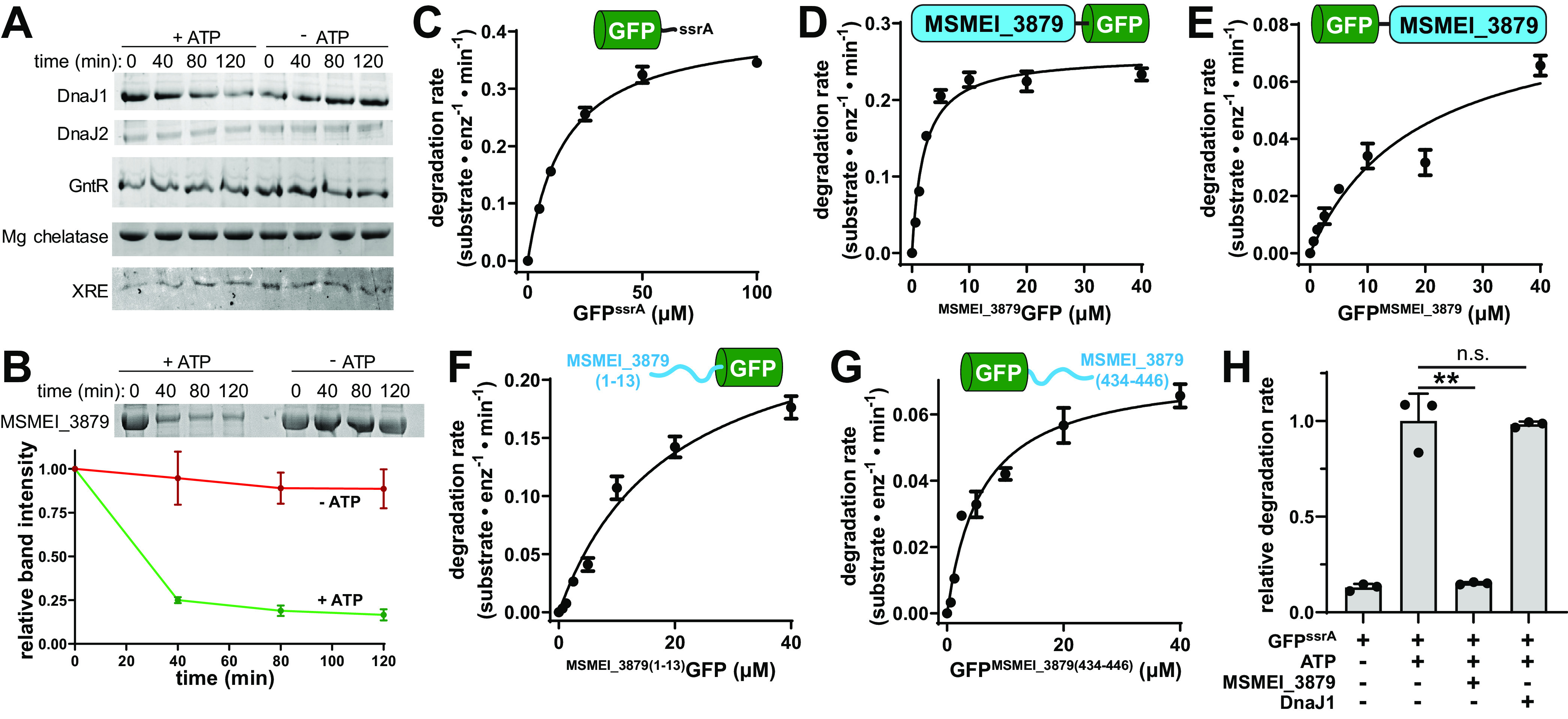
MSMEI_3879 is a substrate of M. smegmatis ClpC1. (A) In vitro degradation assays of DnaJ1, DnaJ2, GntR, magnesium chelatase, and XRE by 1 μM ClpC1 and 1 μM ClpP1P2, monitored by SDS-PAGE. (B) ATP-dependent degradation of 10 μM MSMEI_3879 by ClpC1P1P2 was observed in vitro by SDS-PAGE, with gel densitometry of three replicate assays shown. (C) Michaelis-Menten analysis of GFPssrA proteolysis by 1 μM ClpC1P1P2, as a function of substrate concentration, reveals a kcat of 0.41 ± 0.02 substrate · min−1 · enzyme−1 and a Km of 16.04 ± 1.28 μM. (D) MSMEI_3879GFP was degraded by 1 μM ClpC1P1P2 with a kcat of 0.26 ± 0.01 substrate · min−1 · enzyme−1 and Km of 2.06 ± 0.43 μM. (E) GFPMSMEI_3879 was degraded with a kcat of 0.09 ± 0.02 substrate · min−1 · enzyme−1 and a Km of 18.8 ± 9.6 μM. (F) GFP preceded by the first 13 residues of MSMEI_3879 was degraded with kcat of 0.268 ± 0.033 substrate · min−1 · enzyme−1 and Km of 19.1 ± 4.9 μM. (G) GFP appended with the last 13 residues of MSMEI_3879 was degraded with a kcat of 0.0737 ± 0.0053 substrate · min−1 · enzyme−1 and Km of 5.97 ± 1.3 μM. (H) Degradation of 10 μM GFPssrA by 1 μM ClpC1P1P2 was monitored in the presence and absence of ATP, 10 μM MSMEI_3879, or 10 μM DnaJ1. MSMEI_3879 reduces GFPssrA degradation to the level observed in the absence of ATP. DnaJ1 does not significantly alter the rate of GFPssrA proteolysis. Values are averages of three replicates (n = 3) ± 1 SD. P values were calculated by unpaired two-tailed Student's t test. **, P < 0.01; n.s., not significant.
