ABSTRACT
Arthrobotrys flagrans (Duddingtonia flagrans) is a typical nematode-trapping fungus which has been used for nematode biocontrol. The global regulator LaeA is widely distributed in filamentous fungi and plays a crucial role in secondary metabolism and development in addition to pathogenicity in fungal pathogens. In this study, the chromosome-level genome of A. flagrans CBS 565.50 was sequenced and homologous sequences of LaeA were identified in A. flagrans. A. flagrans LaeA (AfLaeA) knockout resulted in slower hyphal growth and a smoother hyphal surface. Importantly, deletion of AfLaeA resulted in the absence of chlamydospores and attenuated glycogen and lipid accumulation in hyphae. Similarly, disruption of the AfLaeA gene led to fewer traps and electron-dense bodies, lower protease activity, and a delay in capturing nematodes. The AfLaeA gene had a large effect on the secondary metabolism of A. flagrans, and both the deletion and overexpression of AfLaeA could yield new compounds, whereas some compounds were lost due to the absence of the AfLaeA. Protein-protein interactions between AfLaeA and another eight proteins were detected. Furthermore, transcriptome data analysis showed that 17.77% and 35.51% of the genes were influenced by the AfLaeA gene on days 3 and 7, respectively. AfLaeA gene deletion resulted in the higher expression level of the artA gene cluster, and multiple differentially expressed genes involved in glycogen and lipid synthesis and metabolism showed opposite expression patterns in wild-type and ΔAfLaeA strains. In summary, our results provide novel insights into the functions of AfLaeA in mycelial growth, chlamydospore production, pathogenicity, secondary metabolism, and energy metabolism in A. flagrans.
IMPORTANCE The regulation of biological functions, such as the secondary metabolism, development, and pathogenicity of LaeA, has been reported in multiple fungi. But to date, no study on LaeA in nematode-trapping fungi has been reported. Moreover, it has not been investigated whether or not LaeA is involved in energy metabolism and chlamydospore formation has not been investigated. Especially in the formation mechanism of chlamydospores, several transcription factors and signaling pathways are involved in the production of chlamydospores, but the mechanism of chlamydospore formation from an epigenetic perspective has not been revealed. Concurrently, an understanding of protein-protein interactions will provide a broader perspective on the regulatory mechanism of AfLaeA in A. flagrans. This finding is critical for understanding the regulatory role of AfLaeA in the biocontrol fungus A. flagrans and establishes a foundation for developing high-efficiency nematode biocontrol agents.
KEYWORDS: nematode-trapping fungi, Arthrobotrys flagrans, genome, AfLaeA, chlamydospore, pathogenicity, metabolism
INTRODUCTION
The growth, secondary metabolism, and sexual and asexual development of filamentous fungi are regulated by genetic regulators (1). LaeA (loss of aflR expression A) is a global regulator genetically identified from Aspergillus nidulans (2) and is now known to be a phylogenetically conserved methyltransferase in multiple filamentous fungi species (3). Generally, an S-adenosylmethionine (SAM) binding site at the N terminus and a methyltransferase domain at the middle of the LaeA protein are present. Moreover, LaeA protein has a classical nuclear localization signal, which localizes into the nucleus (2). In A. nidulans, LaeA regulates secondary metabolism and fungal development by forming a trimeric protein complex with VeA and VelB in response to light (1). Similarly, Penicillium oxalicum LaeA (PoLaeA) forms a complex with three proteins (Tup1, Cyc8, and PoClrB) to modify chromatin structure in the upstream region of cellulose degradation genes, thereby activating the expression of the cellulose degradation gene in Penicillium oxalicum (3). In more detail, LaeA regulates the expression of cexA via methylation levels of the histones H3K4 and H3K9 in Aspergillus luchuensis (4).
Initially, most of the research on LaeA has been performed in Aspergillus spp., where LaeA is involved in the production of multiple secondary metabolites such as sterigmatocystin, penicillin, and lovastatin (2). Subsequent work revealed that LaeA regulates the production of secondary metabolites of Penicillium spp. (5, 6), Trichoderma spp. (7, 8), the endophytic fungus Epichloe festucae (9), and multiple pathogenic fungi, such as Dothistroma septosporum (10), Alternaria alternata (11), Fusarium oxysporum (12), Fusarium verticillioides (13), and Magnaporthe oryzae (14). The secondary metabolites of pathogenic fungi are closely related to their pathogenicity, and the pathogenicity of multiple pathogenic fungi is also positively regulated by LaeA. For example, deletion of the VmLaeA gene was found to lead to a significant reduction in the virulence of Valsa mali for tobacco (15). Similarly, loss of the LaeA gene in A. alternata (16) and Cochliobolus heterostrophus (17) resulted in a reduced ability to infect tomato and maize leaves, respectively. In addition, the PeLaeA gene mutant strain from Penicillium expansum failed to colonize on apples, and patulin was not detected in apples infected with the mutant strain (18).
LaeA not only regulates the synthesis of fungal secondary metabolites and virulence but also has key functions in fungal development, including the formation of asexual spore and sexual fruiting bodies (1, 14). In A. nidulans, the absence of AnLaeA results in the lack of Hülle cells, a specific globose cell type, which nurse the young fruiting body during development. Because of the absence of this gene, fruiting bodies formed in AnLaeA mutant strains are smaller (19, 20). Similarly, loss of MrLaeA results in the lack of ascospores in Monascus ruber (21). In addition, LaeA is essential to support asexual spore formation in A. nidulans (19, 20), Aspergillus pachycristatus (22), and Aspergillus carbonarius (23), and the number of asexual spores in LaeA mutant strains showed a significant decrease in the light. Apart from Aspergillus spp., LaeA is evidently essential for sporulation in other filamentous fungi. LaeA positively regulates the conidial production of Trichoderma spp., including Trichoderma longibrachiatum SMF2Shi, T. atroviride, and T. reesei (24, 25), Penicillium spp., including P. oxalicum, P. chrysogenum, and P. expansum (26–28), Chaetomium globosum (29), A. alternata (16), and Ganoderma lingzhi (30) but negatively regulates the conidial production of C. heterostrophus (17) and M. ruber (21). LaeA is crucial for development in filamentous fungi.
As a major source for the development of biological nematicides, nematode-trapping (NT) fungi capture nematodes by forming specialized trapping devices (traps), such as adhesive networks, knobs, and constricting rings (31). Arthrobotrys flagrans (formerly Duddingtonia flagrans) is a typical NT fungus that is easy to cultivate and has a strong capability to capture nematodes by producing adhesive networks (31, 32). Importantly, the capability of A. flagrans to produce a large number of chlamydospores is a unique advantage over other NT fungi (33). Biocontrol agents developed from this strain have been successfully used to control parasitic nematodes in multiple animals such as cattle, sheep, horses, chickens, and pigs, among others (34). It is believed that A. flagrans has the potential for development as a biocontrol agent for plant-parasitic nematodes, such as Meloidogyne spp. and Xiphinema index (32–35). LaeA is widespread in filamentous fungi (14, 36), whereas its function remains unclear in NT fungi.
In this study, based on chromosome-level genome sequencing, we identified AfLaeA in the NT fungus A. flagrans. The effects of AfLaeA on hyphal growth, chlamydospore production, virulence, and secondary and energy metabolic processes were characterized by gene knockout, gene complementation, and multiphenotype and transcriptome data analysis.
RESULTS
Chromosome-level genome assembly and annotation of A. flagrans.
The first genome data (scaffold level) of A. flagrans CBS 349.94 were reported in 2019 (32). In the present study, the chromosome-level genome of strain A. flagrans CBS 565.50 was sequenced and analyzed. This study yielded 8.91 Gb of initially filtered PacBio data, and 99.07% of reads were mapped against the genome of A. flagrans CBS 349.94 with 109.58× genome coverage (Table 1). The genome sequence of 35.85 Mb was then assembled using Hifiasm software, and the genome was found to be located on six chromosomes (Fig. 1A), containing six lachesis groups with an N50 scaffold size of 6,108,558 bp and an N90 scaffold size of 4,110,191 bp (Table 1). Importantly, compared with the genome data of A. flagrans CBS 349.94, the gene cluster was found to have more protein-coding genes (up to 9,997) with an average gene size of 1,701.43 bp than found in the A. flagrans CBS 565.50 genome (Table 1). One hundred thirty-three effector proteins and 1,805 transmembrane proteins were also predicted. In addition, the A. flagrans CBS 565.50 genome has 11 secondary metabolism gene clusters, including four polyketide synthase (PKS), two nonribosomal peptide synthase (NRPS), and two terpene gene clusters and one siderophore, one indole, and one betalactone gene cluster (see Fig. S1 and Table S1 in the supplemental material), whereas the genome of A. flagrans CBS 349.94 has only three PKS and three predicted NRPS type gene clusters (32). Furthermore, 733 secreted proteins (secretome) were predicted in the A. flagrans CBS 565.50 genome, whereas 638 secreted proteins were predicted in the A. flagrans CBS 349.94 genome (32). Among these secreted proteins, small secreted proteins (SSPs) have been implicated in fungal pathogenicity (32), and the small secreted cysteine-rich protein CyrA was confirmed as a virulence factor participating in the attack of Caenorhabditis elegans in A. flagrans (37).
TABLE 1.
Features of the chromosome-level genome of A. flagrans CBS 565.50
| Sequencing feature | Valuea |
|---|---|
| Genome size (Mb) | 35.85 |
| Chromosomes | 6 |
| Clean data (Gb) | 8.91 |
| Depth (×) | 109.58 |
| Mapped (%) | 99.07 |
| Coverage (%) | 99.96 |
| Lachesis groups | 7 |
| N50 read length (bp) | 6,108,558 |
| Max contig length (bp) | 8,301,709 |
| N90 length of scaffolds (bp) | 4,110,191 |
| % ≥ Q30 | 92.84 |
| %G+C | 45.57 |
| Repeat content (%) | 5.16 |
| tRNA genes | 177 |
| rRNA genes | 98 |
| Total length of coding sequences (bp) | 17,009,220 |
| Avg gene size (bp) | 1,701.43 |
| Avg no. of exons/gene | 3.61 |
| Avg no. of introns/gene | 2.61 |
| Avg CDS size (codons) | 403.46 |
| Avg intron length (bp) | 94.06 |
| Protein-encoding genes | 9,997 |
| Gene clusters | 11 |
| No. (%) of secretomes | 733 (~7.33) |
| Transmembrane proteins | 1,805 |
| Effector proteins | 133 |
Values are the numbers of the indicated feature unless otherwise specified.
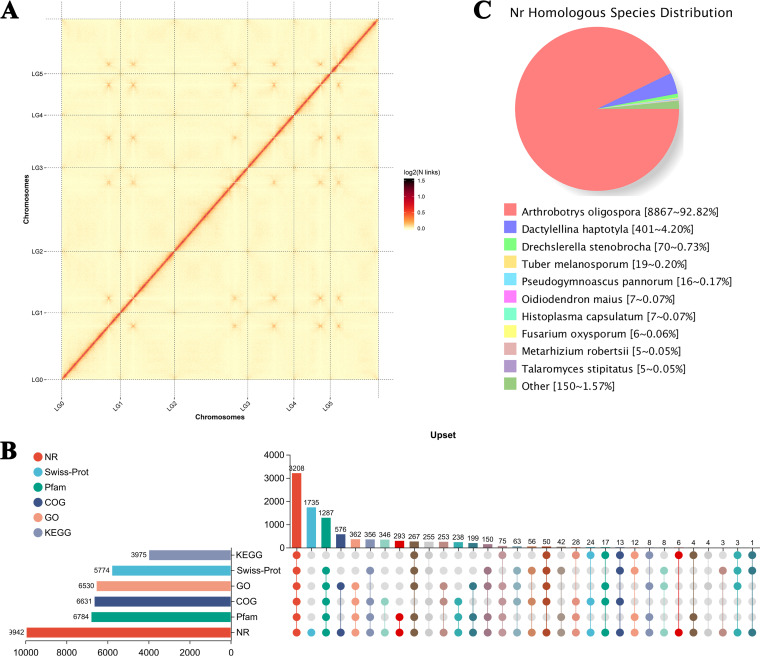
FIG 1.
Chromosome-level genome assembly and annotation of Arthrobotrys flagrans. (A) Interactive heat map of Hi-C assembled chromosomes. LG00 to LG05 represent lachesis groups 00 to 05, and the abscissa and ordinate represent the order of each bin on the corresponding chromosome group. (B) The UpSet diagram of gene annotation statistics. The horizontal histogram on the left shows the statistical values of elements of each set. In the middle matrix, single points represent the unique elements of a set, the connections between points represent the unique intersections of different sets, and the vertical histogram represents the corresponding intersection element values, respectively. (C) Species distribution map of protein sequences compared with the Nonredundant Protein (NR) database.
To further understand gene function in A. flagrans, 6,530, 3,979, 6,631, 9,942, 5,774, and 6,784 proteins were successfully assigned to their orthologs in the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Clusters of Orthologous Genes (COG), Nonredundant Proteins (NR), Swiss-Prot, and Pfam databases, respectively (Fig. 1B). There were 3,208 proteins that could be annotated in the six databases, and only 1,735 genes were found to belong to the NR database (Fig. 1B). Likewise, according to the comparison results of the NR database, A. flagrans and Arthrobotrys oligospora have the highest similarity, with 8,867 homologous sequences, and A. flagrans has 401, 70, 19, 16, 7, 7, 6, 5, and 5 sequences homologous with Dactylellina haptotyla, Drechslerella stenobrocha, Tuber melanosporum, Pseudogymnoascus pannorum, Oidiodendron maius, Histoplasma capsulatum, F. oxysporum, Metarhizium robertsii, and Talaromyces stipitatus, respectively (Fig. 1C).
Identification and sequence analyses of AfLaeA and nine methyltransferases in A. flagrans.
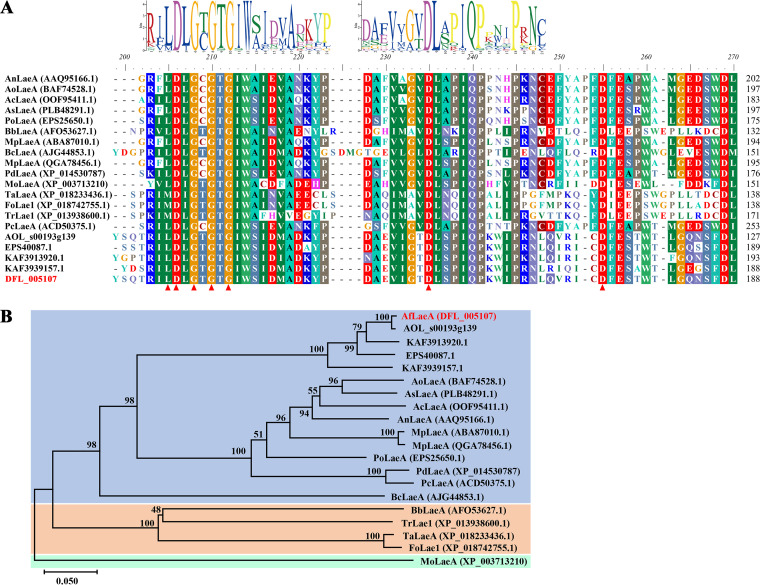
Based on genomic data, the amino acid sequence of the A. nidulans global regulator AnLaeA (GenBank accession number AAQ95166.1) (1) was used for a BLAST search of the A. flagrans genome using National Center for Biotechnology and Information (NCBI) and UniProt software, and one AfLaeA protein (DFL_005107; GenBank accession number RVD86853.1) and nine methyltransferases were predicted, all of which contain a conserved SAM binding site (Fig. 2A; Table S2). AfLaeA and AnLaeA had the highest similarity, with 48.47% (Fig. 2A; Table S3), while the similarities of other methyltransferases were less than 37.92% (Table S3). The AfLaeA gene has seven exons and six introns, and the coding sequence (CDS) contains 1,518 bp that encodes 362 amino acids with a molecular weight of 41.491 kDa (Table S4). The subcellular localization prediction analysis revealed that AfLaeA and AnLaeA were localized to the cytoplasm and nucleus (Table S4). In addition, the homologous protein sequences of LaeA were also found in the nematode-trapping fungi A. oligospora, D. haptotyla, D. brochopaga, and Dactylella cylindrospora (Fig. 2A). They were clustered with AnLaeA in the phylogenetic tree (Fig. 2B). LaeA proteins from different species in the phylogenetic tree have been reported to be involved in biological processes such as fungal growth and development, conidial production, and secondary metabolism (Fig. 2).
FIG 2.
Conserved domains and phylogenetic analysis of LaeA in different fungi. (A) Homologous sequences of LaeA in different fungi were retrieved from the NCBI database, and the conserved domains were analyzed using BioEdit software. The GenBank accession numbers of the homologous sequences of LaeA in nematode-trapping fungi A. oligospora, D. haptotyla, D. brochopaga, and D. cylindrospora are XP_011127052.1, EPS40087.1, KAF3913920.1, and KAF3939157.1, respectively. The GenBank accession numbers of these LaeA proteins are shown in the figure. Red triangles indicate a conserved SAM binding site, and the logo of the conserved domain was predicted using MEME online software. (B) Phylogenetic tree constructed by MEGA X software using the neighbor-joining method. Bootstrap values based on 1,000 replicates are shown in the phylogenetic tree, and AfLaeA (DFL_005107) is highlighted in red.
Deletion of AfLaeA affects fungal growth, morphology, and stress response in A. flagrans.
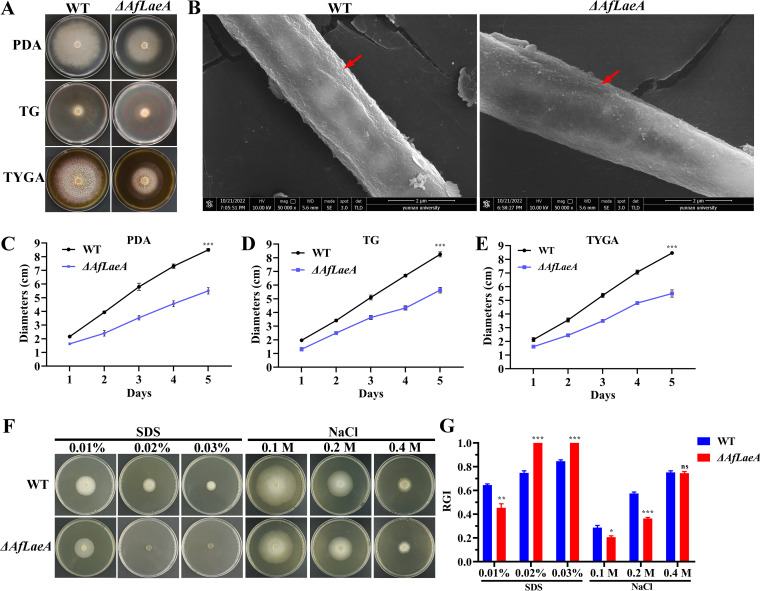
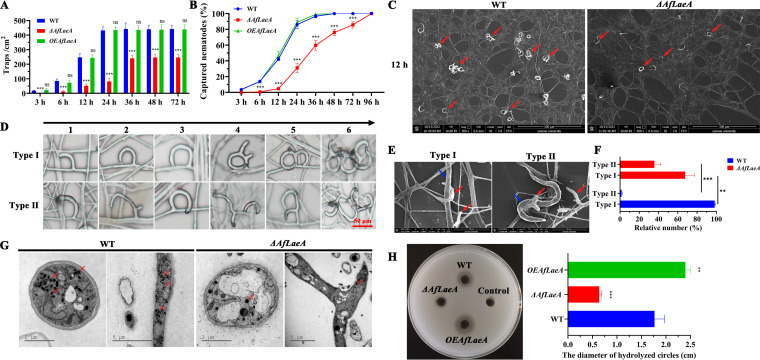
In this study, we deleted the AfLaeA gene by a homologous recombination method to investigate its function in A. flagrans (Fig. S2). Likewise, three methyltransferase genes (DFL_000451, DFL_006623, and DFL_007594) were knocked out in the same way (Fig. S3). Deletion of AfLaeA gene resulted in significantly slower hyphal growth on potato dextrose agar (PDA), TG (1% tryptone, 1% glucose, 1.5% agar), and TYGA (1% tryptone, 0.5% yeast extract, 1% glucose, 0.5% molasses, 1.5% agar) media, with colony diameters of approximately 60% of that of the wild-type (WT) strain on day 5 (Fig. 3A and C to E). Complementation of AfLaeA restored colony growth (Fig. S5A and B). However, the loss of three methyltransferase genes (DFL_000451, DFL_006623, and DFL_007594) had no effect on hyphal growth (Fig. S4). Moreover, the AfLaeA gene was overexpressed in the WT strain, its expression was upregulated 3.5-fold (Fig. S5D and E), and AfLaeA was localized to both the nucleus and cytoplasm (Fig. S5F), which was consistent with the predicted results based on the Cell-PLoc software (Table S4). In addition, scanning electron microscopy (SEM) results showed that the surface of the ΔAfLaeA strain lacked wrinkles in comparison with the WT (Fig. 3B).
FIG 3.
Comparison of growth, morphologies, and stress responses between WT and ΔAfLaeA strains. (A) Colony morphologies of the WT and ΔAfLaeA strains incubated on PDA, TG, and TYGA plates at 28°C for 5 days. (B) Analysis of the cell surface morphology of WT and ΔAfLaeA strains by SEM. Red arrows indicate wrinkled areas on the hyphal surface. (C to E) Colony diameters of the WT and ΔAfLaeA strains cultured on PDA, TG, and TYGA plates at 28°C for 5 days (***, P < 0.001). (F) Growth of the WT and ΔAfLaeA strains on medium supplemented with SDS at 0.01% to 0.03% and NaCl (0.1 M, 0.2 M, and 0.4 M). (G) RGI values of the WT and ΔAfLaeA strains on medium supplemented with SDS (0.01% to 0.03%) and NaCl (0.1, 0.2, and 0.4 M) (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
To confirm the effect of AfLaeA gene on stress resistance of A. flagrans, the osmotic pressure stress reagent NaCl (0.1, 0.2, and 0.3 M) and the cell wall stress reagent sodium dodecyl sulfate (SDS) at 0.01% to 0.03% were added to the PDA medium (Fig. 3F). The results showed that the growth of the WT and ΔAfLaeA strains could be significantly inhibited by 0.01% SDS and the growth of the ΔAfLaeA strain could be completely inhibited by 0.02% SDS (Fig. 3F). The ΔAfLaeA strain was more sensitive to SDS, with higher relative growth inhibition (RGI) values than those of the WT strain (Fig. 3F and G). Similarly, the growth of the WT and ΔAfLaeA strains could be inhibited by 0.1 M NaCl, and the WT strain displayed greater sensitivity to 0.1 M NaCl, with higher RGI values than those of the ΔAfLaeA strain (Fig. 3F and G). Interestingly, the WT and ΔAfLaeA strains had the same sensitivity to 0.4 M NaCl with the same RGI values (Fig. 3G).
AfLaeA plays a key role in chlamydospore production in A. flagrans.
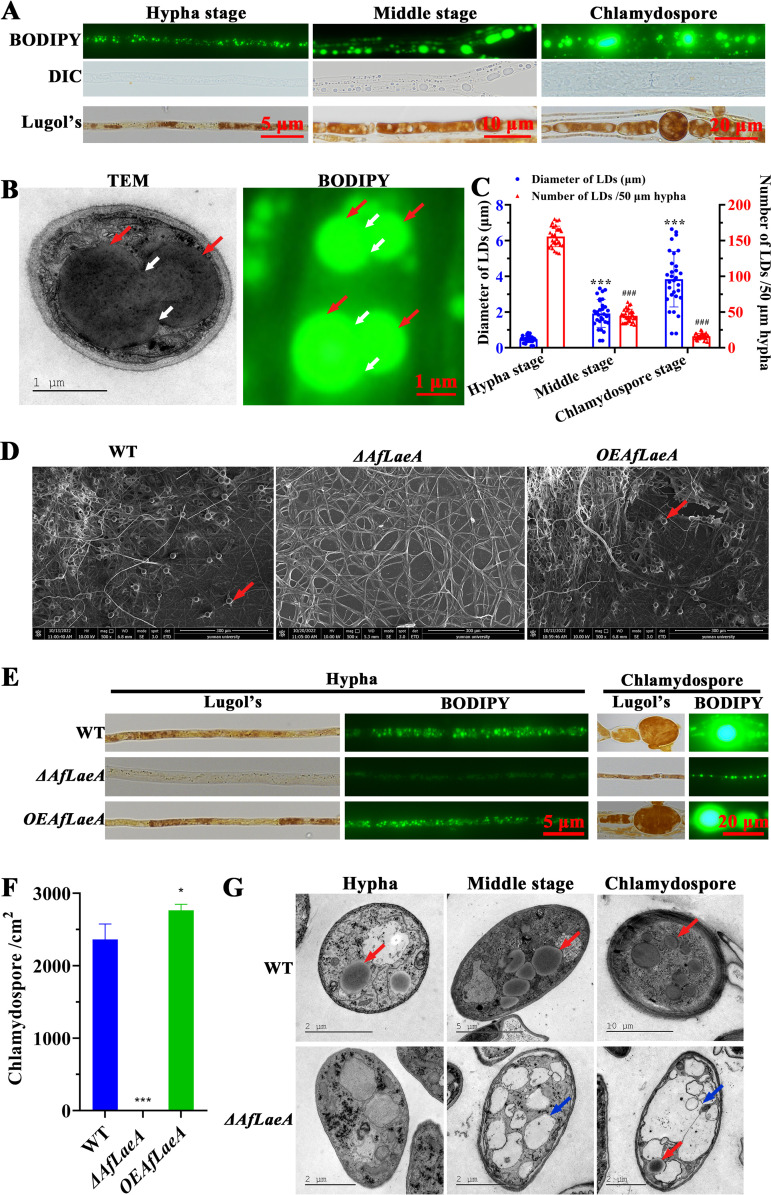
In previous reports, LaeA was involved in the formation of conidia in several fungi (19–29), but it has not been reported to regulate chlamydospore formation. The chlamydospore is a prominent feature of A. flagrans strains. The strong stress resistance of chlamydospore has great significance for the biological control of nematodes. In this study, we observed the distribution of glycogen and lipid droplets (LDs) in hyphae and chlamydospores. In the hyphal stage, LDs in the cells were small and dispersed, but the volume was increased and the distribution was concentrated in the middle stage of chlamydospore production and in the chlamydospore stage (Fig. 4A). Importantly, the fusion phenomenon of LDs was observed by transmission electron microscopy (TEM) and BODIPY staining analysis (Fig. 4B). At the same time, the diameters of the LDs increased but the numbers of LDs decreased during formation of chlamydospores (Fig. 4C). This fully showed that the larger LDs were derived from the fusion of smaller LDs. Similarly, glycogen was also inappropriately increased during chlamydospore production, although we did not observe very significant fusion (Fig. 4A).
FIG 4.
AfLaeA affects the formation of glycogen, LDs, and chlamydospores. (A) Staining analysis of glycogen and LDs at different stages (hyphal stage, middle stage of chlamydospore production, and chlamydospore stage) of chlamydospore formation. The LDs and glycogen were stained with BODIPY and Lugol’s iodine, respectively. DIC, differential interference contrast. (B) Observation of the fusion phenomenon of LDs by TEM and BODIPY staining. Red and white arrows indicate LDs and the gaps caused by the fusion of LDs, respectively. (C) Comparison of the diameters and numbers of the LDs in the process of chlamydospore formation. The diameters of LDs were collected from TEM images of more than 30 cells, and the number of LDs was obtained from a random hypha (50 μm) in more than 30 fields viewed under a microscope. The asterisks and pound signs indicate a significant difference between the hyphal stage and middle stage of chlamydospore production and the chlamydospore stage (Tukey’s honestly significant difference [HSD]; ***, P < 0.001; ###, P < 0.001). (D) Observation of the chlamydospores in WT, ΔAfLaeA, and OEAfLaeA strains using SEM. Red arrows indicate chlamydospores. (E) Comparison of the glycogen and LDs in WT, ΔAfLaeA, and OEAfLaeA strains in hyphal and chlamydospore stages, respectively. (F) Numbers of chlamydospores in WT, ΔAfLaeA, and OEAfLaeA strains (*, P < 0.05). (G) TEM analysis of the internal structure in WT and ΔAfLaeA strains at the hyphal and middle stages of chlamydospore production and the chlamydospore stage, respectively. Red and blue arrows indicate LDs and vacuoles, respectively.
Importantly, deletion of AfLaeA gene resulted in the absence of chlamydospores, and this defect could be restored by complementation in the ΔAfLaeA strain (Fig. S5C). Interestingly, the overexpression (OE) of AfLaeA produced chlamydospores and increased their production by approximately 16% (Fig. 4D to F). In addition, the ΔAfLaeA strain showed defects in glycogen and LD formation and accumulation, although more glycogen and LDs were formed at the end of hyphal growth (Fig. 4E). Additionally, the accumulation of glycogen and LDs was not significantly increased in overexpressed AfLaeA (OEAfLaeA) strains (Fig. 4E). Of note, the deletion of a methyltransferase gene (DFL_000451) resulted in a 30% reduction in chlamydospores but had no significant effects on the accumulation of glycogen and LDs (Fig. S6). Furthermore, the accumulation of glycogen and LDs during chlamydospore formation in the WT strain was also observed using TEM, as shown in Fig. 4G. We also found that the loss of the AfLaeA gene resulted in the appearance of numerous vacuoles in cells at the same stage (Fig. 4G).
Effect of AfLaeA on pathogenicity of A. flagrans.
As an NT fungus, the pathogenicity of A. flagrans for nematodes is an important function (38, 39). In this study, the effects of the AfLaeA gene on the production of traps and the capability to capture nematodes were investigated. The results showed that deletion of AfLaeA gene resulted in significantly fewer traps and a lower production rate of traps (Fig. 5A and C). After the addition of nematodes for 24 h, the WT strain could produce approximately 430 traps/cm2, twice as many as produced by the ΔAfLaeA strains (Fig. 5A). The traps of the WT strain became saturated after 24 h, whereas ΔAfLaeA strains required 36 h (Fig. 5A). Similarly, ΔAfLaeA strains required 96 h to capture all nematodes, whereas the WT strain required only 36 h (Fig. 5B). The WT strain produced two types of traps (regular and irregular) upon induction by nematodes (Fig. 5D), and both types of traps could catch and kill nematodes (Fig. 5E). Interestingly, the loss of the AfLaeA gene resulted in the appearance of more irregular (type II) traps, up to 34%, whereas about 1% of this type of trap was found in the WT strain (Fig. 5F). In addition, electron-dense bodies (EDs) in the trap cells of the WT and ΔAfLaeA strains were observed using TEM, and the results showed that fewer EDs were observed in the ΔAfLaeA strains than in the WT (Fig. 5G). Extracellular protease activity is also an indicator of fungal pathogenicity (38). Deletion of AfLaeA gene resulted in a significant decrease in extracellular protease activity, whereas overexpression of the AfLaeA gene resulted in enhanced activity (Fig. 5H). The WT strain showed approximately 3-fold-higher extracellular protease activity than the ΔAfLaeA strain, whereas the OEAfLaeA strains showed 1.5-fold-higher activity than the WT strain (Fig. 5H).
FIG 5.
Comparison of trap formation, trap morphologies, nematocidal activities, EDs, and extracellular proteolytic activities. (A) Comparison of traps produced by WT, ΔAfLaeA, and OEAfLaeA strains at different time points (nonsignificant [ns], P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001). (B) Percentage of captured nematodes at different time points. (C) SEM analysis of the traps at 12 h. Red arrows indicate traps. (D) Comparison of the trap morphologies in WT and ΔAfLaeA strains. The numbers at the top of the panel (1 to 6) represent the different stage of trap formation. (E) Nematodes were captured by two types of traps. Red and blue arrows indicate nematodes and traps, respectively. (F) Percentage of two types of traps in WT and ΔAfLaeA strains (***, P < 0.001). (G) Comparison of EDs in trap cells of WT and ΔAfLaeA strains based on TEM. Red arrows indicate EDs in trap cells of WT and ΔAfLaeA strains. (H) Comparison of extracellular protease activities (**, P < 0.01; ***, P < 0.001).
Prediction and analysis of interactional proteins of AfLaeA.
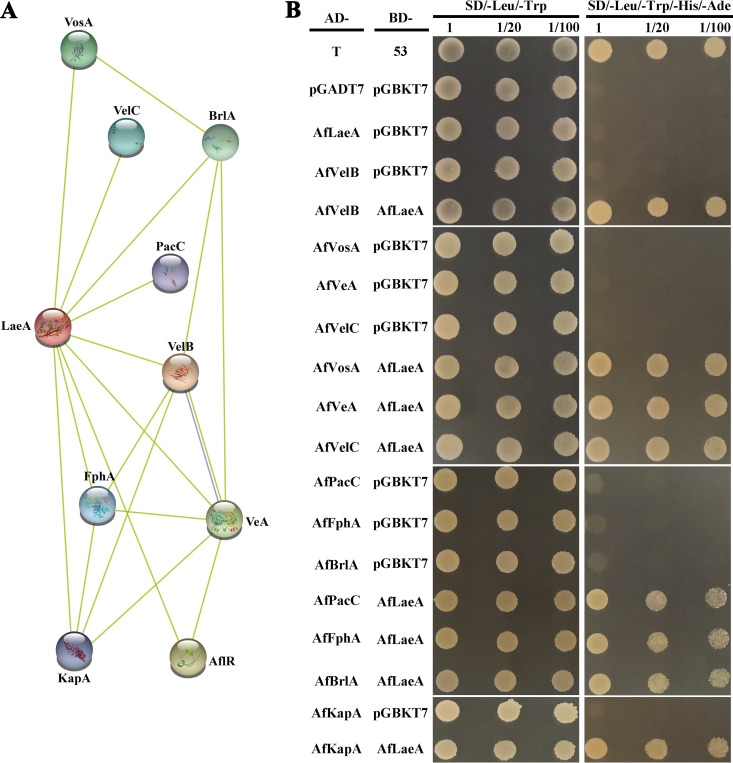
LaeA, as a methyltransferase, must interact with other proteins to form complexes, thereby regulating biological processes such as light response, secondary metabolism, growth, and development (1, 2, 11). In this study, we used AnLaeA as the query sequence and STRING online software to predict the interaction protein of LaeA. The proteins that interacted with AnLaeA included four velvet family proteins (VosA, VeA, VelB, and VelC), pH response transcription factor PacC, phytochrome FphA, C2H2 type master regulator BrlA, importin subunit alpha KapA, and sterigmatocystin biosynthesis regulatory protein AflR (Fig. 6A). With the exception of AflR, the other eight interaction proteins in A. flagrans could be predicted (Fig. 6B). Subsequently, this prediction was verified using the yeast two-hybrid (Y2H) assay, which showed that AfLaeA interacted with four velvet family proteins (AfVosA, AfVeA, AfVelB, and AfVelC), AfPacC, AfFphA, AfBrlA, and AfKapA (Fig. 6B). This result provided a good foundation for the subsequent experiments.
FIG 6.
Analysis of the interacting protein with AfLaeA in the yeast system. (A) Prediction of LaeA-interacting networks using STRING software (http://string-db.org/). (B) Y2H analysis of the interactions between AfLaeA and four velvet family proteins (AfVosA, AfVeA, AfVelB, and AfVelC), AfPacC, AfFphA, AfBrlA, and AfKapA.
Transcriptomic profile analysis of the WT and AfLaeA strains.
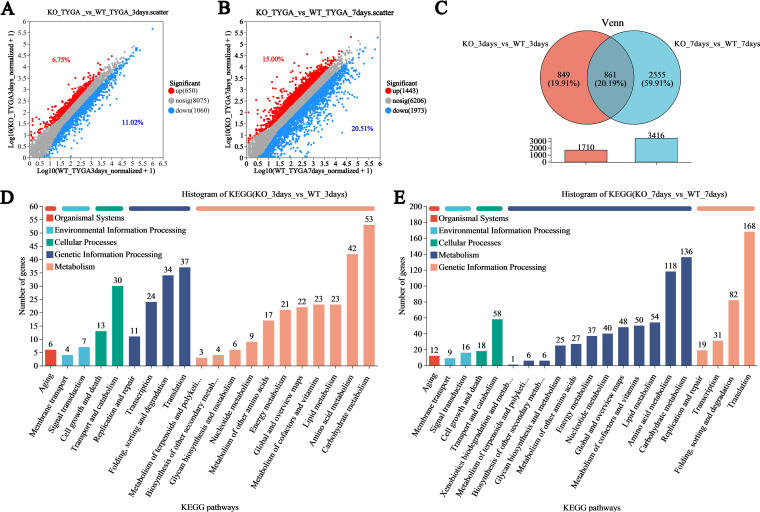
To further investigate the regulatory mechanisms of AfLaeA in A. flagrans, the transcriptomic profiles of the WT and ΔAfLaeA strains were compared using transcriptome sequencing (RNA-seq). The results showed that the number of clean reads that could be located on the genome in each sample was more than 95% relative to that of the genome (Table S5). Principal-component analysis (PCA) showed that the WT and ΔAfLaeA strains were located in different quadrants at various time points, and the three duplicate samples had high similarity (Fig. S7). ΔAfLaeA strains were found to have 1,710 and 3,416 differentially expressed genes (DEGs) at 3 and 7 days, respectively (Fig. 7A and B). The number of downregulated and upregulated DEGs accounted for 11.20% and 6.75% and 20.51% and 15.00% at 3 days and 7 days, respectively (Fig. 7A and B). In addition, 849 and 2,555 DEGs were specifically expressed at 3 and 7 days, respectively (Fig. 7C).
FIG 7.
Transcriptomic insight into the regulatory role of AfLaeA (A and B) Differentially expressed genes (DEGs) on days 3 and 7, respectively. (C) Venn analysis of the DEGs at two time points in WT and ΔAfLaeA strains. (D and E) KEGG enrichment analysis of DEGs in WT and ΔAfLaeA strains.
On the third day, GO enrichment analysis showed that the GO terms most enriched by upregulated DEGs in the ΔAfLaeA strain were associated with the preribosome, nucleolus, and rRNA metabolic process, among others, whereas the downregulated DEGs were associated with the integral component of the membrane, the extracellular region, and the anchored component of the membrane, among others (Fig. S8A and B). On day 7, the upregulated GO terms included mitochondrial transport, tRNA aminoacylation for protein translation, and translational elongation, among others (Fig. S8C). Additionally, the downregulated GO terms included carbohydrate metabolic process, hydrolase activity, and DNA-binding transcription factor activity, among others (Fig. S8D). KEGG analysis showed that more DEGs were present in KEGG pathways on day 7, but the KEGG pathway types enriched for upregulated and downregulated genes, including lipid metabolism, energy metabolism, carbohydrate metabolism, aging, glycan biosynthesis and metabolism, cell growth and death, global and overview maps, signal transduction, folding, metabolism of terpenoids and polyketides, biosynthesis of other secondary metabolites, and nucleotide metabolism, among others, were the same on days 3 and 7 (Fig. 7D and E).
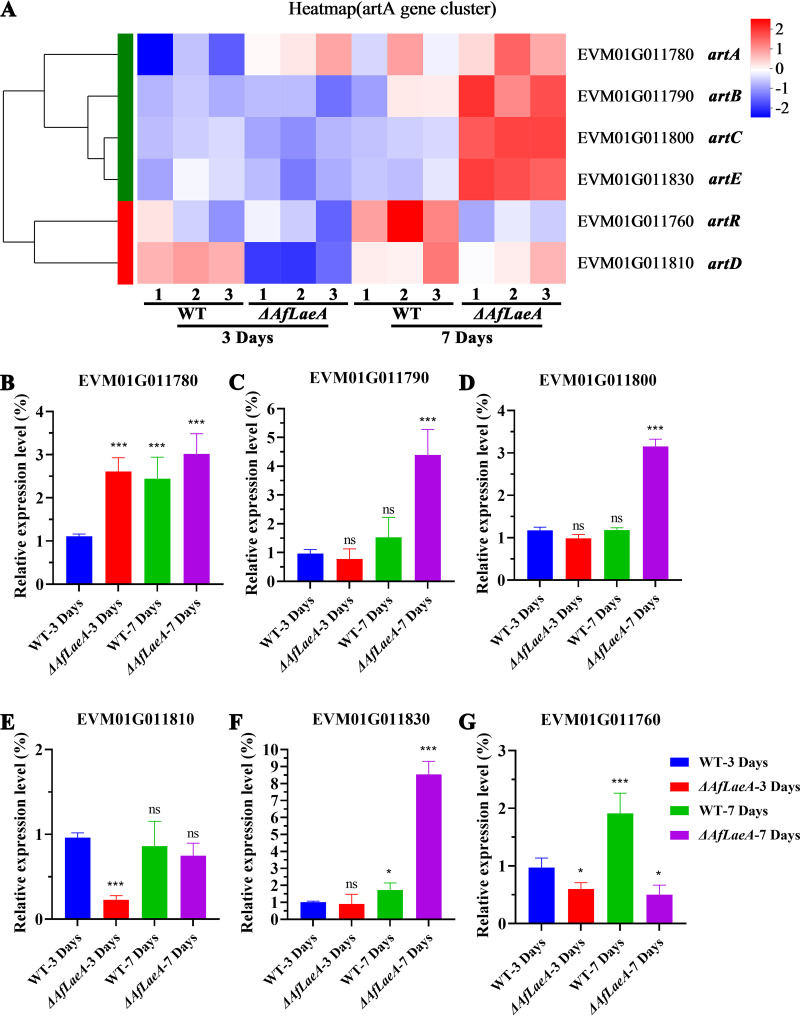
On the basis of the GO and KEGG analyses, we analyzed the artA gene cluster, which plays a key role in A. flagrans and A. oligospora (Fig. S9). The 6-methylsalicylic acid (6-MSA) produced by the artA gene cluster can fatally attract nematodes and regulate the production of traps in time and space (39). In this study, we found that the expression level of most genes in the artA gene cluster was low on day 3, but their expression level increased on day 7 (Fig. 8). The artA and artD genes had opposite expression patterns on day 3, but the patterns tended to converge by day 7 in the WT and ΔAfLaeA strains (Fig. 8A). Importantly, the expression level of the artA gene cluster in the ΔAfLaeA strain was found to be significantly higher than that of the WT strain, except for the artR gene (Fig. 8). Furthermore, the same results were obtained by quantitative real-time PCR (qPCR), as shown in Fig. 8B to G.
FIG 8.
AfLaeA affects the expression of the artA gene cluster. (A) Transcriptome analysis of the expression level of the artA gene cluster at days 3 and 7. (B to G) The expression level of the artA gene cluster was analyzed by qPCR (ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
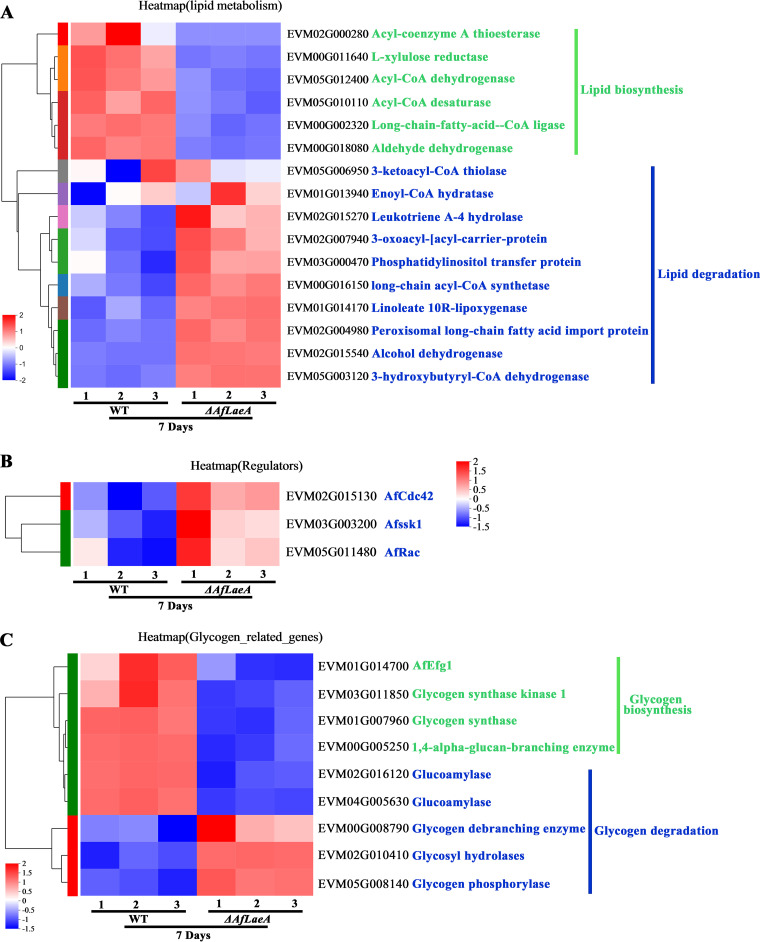
We analyzed several genes that were enriched in relation to lipid and glycogen metabolism. The DEGs involved in lipid synthesis and degradation showed different expression patterns in the WT and ΔAfLaeA strains (Fig. 9). Among the DEGs related to lipid metabolism in the ΔAfLaeA strains, six genes were involved in lipid biosynthesis (genes coding for acyl coenzyme A [acyl-CoA] thioesterase, acyl-CoA dehydrogenase, and acyl-CoA desaturase, among others), and 10 genes were involved in lipid degradation (genes coding for enoyl-CoA hydratase, long-chain acyl-CoA synthetase, and peroxisomal long-chain fatty acid import protein, among others) (Fig. 9A). In addition, the expression patterns of some regulatory genes (AfCdc42, AfSSK1, and AfRac) were also analyzed in the WT and ΔAfLaeA strains (Fig. 9B). The deletion of AoCdc42, AoSSK1, and AoRac genes led to accumulation of LDs in A. oligospora (40, 41). In this study, the deletion of AfLaeA led to upregulation of the expression levels of the AfCdc42, AfSsk1, and AfRac genes (Fig. 9B). Similarly, the effect of AfLaeA on glycogen production showed the same patterns, including glycogen biosynthesis and degradation (Fig. 9C). Glycogenin, glycogen synthase, and 1,4-α-glucan-branching enzymes are required for glycogen synthesis, while glycogen phosphorylase, glycogen debranching enzyme, glucoamylase, and glycosyl hydrolase are required for degradation. The glycogen biosynthesis-related genes were all less strongly expressed in ΔAfLaeA strains, but degradation-related genes were all more strongly expressed (Fig. 9C). In addition, AfEfg1, as a transcription factor that regulates the expression of glycogen synthase (42), also showed a lower expression level in ΔAfLaeA strains (Fig. 9C). Interestingly, two glycogen-degrading enzymes (glucoamylase) have high expression levels in the WT strain (Fig. 9C).
FIG 9.
AfLaeA affects the expression of LDs and glycogen metabolism. (A) Transcriptome analysis of the expression level of LD metabolism genes at days 3 and 7. (B) Transcriptome analysis of the expression level of AfCdc42, AfSSK1, and AfRac at days 3 and 7. (C) Transcriptome analysis of the expression level of glycogen metabolism genes at days 3 and 7.
Secondary metabolism insight into the regulatory role of AfLaeA in A. flagrans.
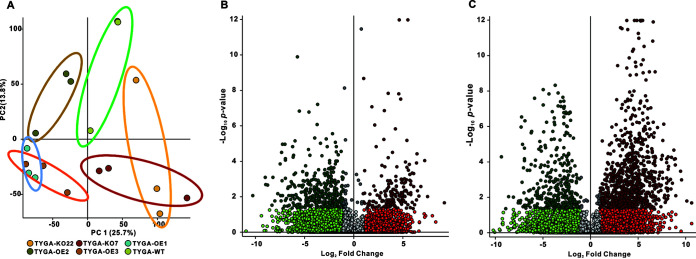
Regulation of secondary metabolism is one of the main functions of LaeA in fungi (6). In this study, to investigate the effect of LaeA on secondary metabolism, the WT, ΔAfLaeA, and OE strains were cultured in TYGA medium for 14 days. Subsequently, we used liquid chromatography-mass spectrometry (LC-MS) and untargeted metabolomics to evaluate the function of AfLaeA involved in secondary metabolite regulation in A. flagrans. PCA showed that the secondary metabolites produced by the WT, ΔAfLaeA, and OE strains were independently distributed in the quadrants (Fig. 10A). Under the screening conditions of a P value of <0.05 and a fold change difference of >2, we observed 171 upregulated and 340 downregulated metabolites in ΔAfLaeA strains (Fig. 10B), while 860 upregulated and 353 downregulated metabolites were found in the OE-3 strain (Fig. 10C). Among these differential metabolites, the most downregulated (564.56-fold) compound was found at an m/z of 331.2378 with a retention time of 17.007 min in ΔAfLaeA strains, while the most upregulated (1,204.756-fold) compound in the OE-3 strain was found at an m/z of 445.2958 with a retention time of 17.186 min (Table S6). The top 20 downregulated and unregulated metabolites of the ΔAfLaeA and OE strains are listed in Table S6. According to the chromatogram, deletion of AfLaeA resulted in the disappearance of several metabolites, but several new compounds appeared (Fig. S10). The major missing secondary metabolites in ΔAfLaeA strains reappeared, and several new secondary metabolites were produced by overexpression of AfLaeA (Fig. S10).
FIG 10.
Comparison of metabolic profiles of fermentation broth from the WT, ΔAfLaeA, and OE strains. (A) PCA of the secondary metabolites produced by WT, ΔAfLaeA, and OE strains cultured in TYGA medium for 14 days. TYGA-WT represents the WT strain. TYGA-KO22 and TYGA-KO7 represent the ΔAfLaeA strains. TYGA-OE1, TYGA-OE2, and TYGA-OE3 represent the OE strains. (B) Volcano plot of differential metabolites in ΔAfLaeA strains compared with the WT strain. The 340 compounds on the left half of the plot were present at significantly higher levels in the WT sample. (C) Volcano plot of differential metabolites in OEAfLaeA strains compared with the WT strain. The 860 compounds on the right half of the plot are present at significantly higher levels in the OE-3 sample.
DISCUSSION
In fungi, several global regulators of secondary metabolism, including mitochondrial complex I, SaraC, and LaeA, have been reported (43, 44). To date, LaeA has been identified mainly in multiple ascomycetes but recently also in basidiomycetes (see Table S7 in the supplemental material). Consistently, LaeA has crucial functions in secondary metabolism and fungal development in most ascomycetes (6, 45) in addition to involvement in the pathogenicity of multiple pathogenic fungi (Table S7). As for the pathogenic fungi of animal/plant-parasitic nematodes, several genes, such as the SofT gene (32), the artA gene cluster (39), and genes coding for small secreted cysteine-rich protein CyrA (37) and STRIPAK (striatin-interacting phosphatase and kinase) component SipC (46), are involved in fungal development and virulence toward nematodes in A. flagrans. However, the function of LaeA remains poorly understood in NT fungi.
LaeA is required for mycelial growth of A. flagrans.
Previous studies have shown that deletion of LaeA gene leads to slow growth of Trichoderma spp. (8), C. heterostrophus (17), Pestalotiopsis microspora (47), and Coprinopsis cinerea (48). In the present study, the colony growth of ΔAfLaeA strains was remarkably smaller than that of the WT strain (Fig. 3). In contrast, this phenotypic change in ΔAfLaeA strains did not coincide with those reported in M. ruber, whose MrLaeA mutants formed abnormal colonies that had abundant aerial hyphae with a higher growth rate than that of the WT (21). Interestingly, loss of AnVelB led to a lower growth rate than that of the WT in A. nidulans (49). In addition, the ΔAoVelB mutants showed significant growth defects on PDA, TYGA, and TG media in terms of A. oligospora (50). Importantly, the different combinations of LaeA and velvet family proteins have different biological functions, such as tight regulation of the morphogenesis of filamentous fungi (1, 19, 20), and these interactions were also confirmed in A. flagrans using Y2H assays (Fig. 6). This finding indicates that LaeA and VelB are required for mycelial growth with the same function and that hyphal growth may be regulated by LaeA and VelB complexes.
LaeA is a key regulator of chlamydospore production in A. flagrans.
A chlamydospore is a thick-walled hypopus produced by fungal hyphae under adverse environmental conditions (low temperature, unfavorable pH, and nutrient deficiency, among others), which can help fungi adapt to harsh environments (51). It has been reported that many fungi, such as the unicellular eukaryotes Candida albicans, Candida dubliniensis, and Cryptococcus laurentii (52–54), filamentous fungi F. oxysporum, Trichoderma harzianum, and Metarhizium anisopliae (55, 56), and the macrofungus Coprinus cinereus (57) can produce chlamydospores during the morphological differentiation stage. In the unicellular eukaryote C. albicans, Hog1, cek1, cla4, cst20, hst7, cph1, cpp1, Gcn4, Gln3, NCR, Gat1, Nrg1, Efg1, RME1, Dpm1, and Dpm3 genes are involved in the formation of chlamydospores (58, 59). In addition, the chlamydospores of F. oxysporum and T. reesei are positively regulated by calcineurin gene CNA1 and chitin synthase genes Chs1_7926 and Chs1_8917, respectively (60, 61). Notably, deletion of the methyltransferase phcR gene in Ralstonia solanacearum leads to an increase in chlamydospores (62). In this study, deletion of AfLaeA gene abolished the capability of A. flagrans to produce chlamydospores (Fig. 4).
A lack of the STRIPAK component SipC results in a reduced number of conidia and chlamydospores, and the morphology of 71.3% chlamydospores in mutants is also greatly expanded (46). SipC, as a highly conservative eukaryotic signal center, is necessary for the asexual development of A. flagrans and plays an important role in the determination of cell fate (46). Interestingly, the STRIPAK complex is required for the regulation of fungal development and the correct expression of the secondary metabolic heterotrimeric VeA-VelB-LaeA complex in A. nidulans (63). In Ustilaginoidea virens, chlamydospore formation is regulated by UvAtg7, UvHOX2, and UvVEA. Loss of UvAtg7 and UvHOX2 genes was found to lead to defects in chlamydospore production (64, 65), and loss of the UvVEA gene led to a decrease in the number of chlamydospores (66). A STRING website prediction and Y2H analysis showed that AfLaeA can interact with AfVeA (Fig. 6), but the biological function of AfVeA in A. flagrans is still unknown. The interaction between LaeA and other proteins can strictly regulate the fungal development of Aspergillus spp. and many other fungi (Table S7). These evidences provide the basis for further understanding of the formation mechanism of chlamydospores.
LaeA affects the energy metabolism in A. flagrans.
Apart from the abolished capacity of chlamydospore production, LaeA affects the energy metabolism of A. flagrans, including glycogen and lipid metabolic processes (Fig. 4E). Lipids and glycogen are the main energy storage material of conidia and chlamydospores (42). These energy substances can be degraded during spore germination to provide the energy needed for fungal growth and initial materials for synthesizing other cellular components (67). In this study, deletion of AfLaeA resulted in a reduction in glycogen and lipid accumulation in mycelium (Fig. 4E). In addition, our transcriptomic analysis demonstrated that 21 and 37 DEGs were annotated to the energy metabolism pathway, whereas 23 and 54 DEGs were annotated to the lipid metabolism pathway, at days 3 and 7, respectively (Fig. 7D and E). The expression levels of genes related to lipid and glycogen synthesis and degradation showed opposite trends in the WT and ΔAfLaeA strains (Fig. 9). In terms of lipid metabolism, several genes involved in lipid degradation were upregulated in ΔAfLaeA strains (Fig. 9), and two genes (those encoding enoyl-CoA hydratase and long-chain acyl-CoA synthetase) are involved in the β-oxidation cycle (Fig. 9A), which constitutes the major pathway of fatty acid degradation in most fungi (37). This result indicates that the degradation of fatty acids is enhanced in ΔAfLaeA strains. In addition, two Rho GTPase genes (Rac and Cdc42) and Ssk1 play an important role in lipid accumulation in A. oligospora (40, 41). In this study, regulatory genes (AfCdc42, AfSsk1, and AfRac) had higher expression levels in ΔAfLaeA strains (Fig. 9B).
In terms of glycogen metabolism, glycogen synthesis is controlled by protein kinase A signaling via the transcription factor CaEfg1 in C. albicans. Loss of CaEfg1 results in a decrease in intracellular glycogen content (42). In this study, loss of the AfLaeA gene resulted in a reduction in glycogen accumulation (Fig. 4E) and a reduction in expression levels of AfEfg1 and glycogen synthase (Fig. 9C). In contrast, loss of glycogen synthase YlGSY1 was found to lead to a decrease in glycogen but an increase in lipid content in oleaginous Yarrowia lipolytica. Glycogen synthesis is a competing pathway for lipid accumulation in oleaginous yeasts (68). In A. flagrans, glycogen accumulation and lipid accumulation occur concurrently (Fig. 4), which also illustrates the similarities and differences of energy metabolism in different species.
AfLaeA strongly regulates the secondary metabolism of A. flagrans.
LaeA regulates the expression of multiple secondary metabolic gene clusters and affects the formation of secondary metabolites. Loss of the LaeA gene hindered the expression of multiple secondary metabolic gene clusters, including kojic acid, cyclopiazonic acid, citrinin, citric acid, and the lovastatin gene cluster (2, 30, 69–71). In Valsa mali, about half (31/60) of the secondary metabolism gene clusters are regulated by VmLaeA (15). Similarly, most PKS and NRPS genes were differentially regulated by Lae1 in T. reesei (72). In this study, our results showed that deletion of AfLaeA gene resulted in upregulation of most of the genes (artA, artB, artC, and artE) in the artA cluster (Fig. 8). It was previously reported that arthrosporols produced by the PKS gene cluster (artA) can inhibit trap production in A. flagrans (39). Interestingly, under induction by nematodes, trap production was much slower in ΔAfLaeA strains, and the number of traps was reduced approximately 50% compared with that in the WT (Fig. 5A). This result may be due to the upregulation of some artA cluster genes.
In addition, it has been commonly observed that the overexpression of LaeA has been successfully used to activate some gene clusters, enhance production of several secondary metabolites, and produce new compounds in fungi (45). Overexpression of LaeA increases the transcription and product formation of genes involved in the kojic acid antitumor compounds terrequinone A, cyclopinionic acid, and tertrimilone (2, 30, 69–72). Moreover, overexpression of LaeA results in the production of the novel compound in Aspergillus versicolor 0312 (73), Chaetomium globosum (29), and Penicillium dipodomyis YJ-11 (74), respectively. In A. flagrans, both deletion and overexpression of AfLaeA gene led to significant changes in secondary metabolites. Deletion of AfLaeA gene led to downregulation of 340 metabolites, while overexpression of AfLaeA led to upregulation of 860 metabolites (Fig. 10). Furthermore, deletion or overexpression of AfLaeA gene resulted in the loss of multiple secondary metabolites in addition to the production of multiple new compounds (Fig. S10).
AfLaeA is associated with the pathogenicity of A. flagrans.
LaeA has been shown to positively regulate pathogenicity toward the host in several pathogenic fungi (14–17). The pathogenicity of these pathogenic fungi was quantified based on the degree of colonization in the host and virulence factors such as mycotoxins and serine proteases (50). However, the pathogenicity of NT fungi is demonstrated by the number of traps, the capability to capture nematodes, and the activity of virulence factors, such as small secreted proteins and serine proteases (37, 50). In the present study, deletion of AfLaeA gene resulted in a delay in trap generation, an increased proportion of irregular traps, and a reduction in the number of traps (Fig. 5). The reason for this phenomenon is estimated to be related to the regulation of fungal growth and the artA cluster by AfLaeA. Additionally, compared with that of the WT strain, the proteolytic activity of ΔAfLaeA strains was weakened but that of OEAfLaeA strains was enhanced (Fig. 5H). It was previously reported that the loss of LaeA resulted in strong underexpression of infection-related proteins in Botrytis cinerea (75), reduction of extracellular proteins in Beauveria bassiana (76), and decreases of extracellular hydrolases in A. flavus (77). After the potential interacting protein VelB of LaeA was disrupted, the extracellular proteolytic activities were reduced, and the transcriptional levels of five serine protease genes were downregulated in A. oligospora (50).
The function of LaeA is epigenetic regulation of downstream genes.
Epigenetic mechanisms control the expression of genes in chromosomal regions by regulating their state. When condensed and tightly packed heterochromatin is formed, genes are silenced, whereas when relaxed euchromatin is formed, genes can be transcribed for function (78). LaeA contains a SAM binding domain and appears to methylate histone proteins differentially to alter the chromatin structure for promoting gene expression (1). In a previous report, the SAM domain of BcLAE1 was shown to be involved in abscisic acid biosynthesis in B. cinerea TB-31 (45). However, in A. flagrans, the key active domain of AfLaeA should not be the SAM domain because deletion of the two methyltransferase genes with the same SAM domain has no effect on mycelial growth or chlamydospore production (Fig. S4 and S6). In Aspergillus spp., the expression of LaeA was negatively regulated by the Ras transduction pathway and cyclic AMP/protein kinase A (cAMP-PKA signaling pathway), which can respond to environmental stimuli (2). In terms of growth in such a scenario, an LaeA-mediated process would trigger the removal of heterochromatic marks, thus allowing the transcription of the downstream genes (79). Similarly, when chlamydospores are produced under unfavorable environmental conditions, the production mechanism of chlamydospores may be regulated via a similar signaling pathway involving LaeA.
MATERIALS AND METHODS
Fungal strains and culture conditions.
A. flagrans CBS 565.50 was stored in the Microbial Library of the Germplasm Bank of Wild Species from Southwest China. A. flagrans was cultivated on PDA (Sigma) medium at 28°C. The mutants (ΔAfLaeA, ΔDFL_000451, ΔDFL_006623, and ΔDFL_007594) were cultivated on PDA medium containing 100 μg/mL hygromycin B. The overexpression strains (OEAfLaeA) were cultivated on PDA medium containing 100 μg/mL hygromycin B and 50 μg/mL nourseothricin.
DNA and RNA isolation and gene cloning.
The total genomic DNA was extracted from 4-day-old cultured fungal hyphae using a HiPure fungal DNA kit II (Magen). Extraction of total RNA and first-strand cDNA synthesis were performed using RNA-easy isolation reagent (Vazyme) and a HiScript III first-strand cDNA synthesis kit (+gDNA wiper) (Vazyme), respectively. Genomic DNA and cDNA were used for PCR amplification of the genes or DNA fragments using specific primers (see Table S8 in the supplemental material). Finally, the PCR products were used for agarose gel electrophoresis and sequencing analysis.
Chromosome-level genome assembly and annotation.
Fifteen micrograms of DNA was used to construct a PacBio Sequel reads library by using a SMRTbell express template prep kit 2.0 (PacBio), and PacBio Sequel II technology was used to sequence the whole genome circular consensus sequencing (CCS). The sequencing runs and assembly of the libraries were performed by Biomarker Technologies (China), and the genome analysis was performed using BMKCloud (https://www.biocloud.net).
Sequence and phylogenetic analysis.
The DNA sequences of AfLaeA and the methyltransferase gene were extracted from the genome data of A. flagrans CBS 565.50. The gene structure of AfLaeA was analyzed using the online software Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/). MEME (Multiple Expectation Maximization for Motif Elicitation; http://meme-suite.org/), SMART (Simple Modular Architecture Research Tool; http://smart.embl.de), and NCBI online tool Conserved Domain Search Service (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) were used to identify and annotate the conserved SAM binding site (2). BioEdit software was used for sequence alignment, and the software package MEGA X (http://www.megasoftware.net/) was used for phylogenetic analysis. NCBI’s Basic Local Alignment Search Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to analyze the similarity of proteins. In addition, the subcellular localization of proteins was predicted by Euk-mPLoc 2.0 software (http://www.csbio.sjtu.edu.cn/bioinf/euk-multi-2/).
Plasmid construction.
The AfLaeA gene was knocked out by the homologous recombination method. In brief, two fragments of 1,036 bp and 960 bp corresponding to the 5′ and 3′ regions of the AfLaeA gene, respectively, were amplified using PCR with primer sets ko5107-5-for/rev and ko5107-3-for/rev (Table S8), respectively, from A. flagrans genomic DNA. The hygromycin B resistance cassette (hph) was amplified using PCR with primers (Table S8) from the pCSN44 vector. All fragments were then assembled in the pUC19 vector (digested with EcoRI and NdeI) by using the ClonExpress Ultra one-step cloning kit (Vazyme). Subsequently, the gene knockout fragment of the AfLaeA gene was amplified by PCR with primers ko5107-5-for and ko5107-3-rev and recovered to a concentration of 5 to 10 μg/μL. The gene knockout plasmid of three methyltransferase genes (DFL_000451, DFL_006623, and DFL_007594) were assembled by the same method (Fig. S2).
For overexpression, 2,000- and 600-bp PCR products containing the promoter and downstream regions of the glyceraldehyde-3-phosphate dehydrogenase (AfGpd) gene, respectively, were amplified using the specific primers (Table S8). The 720-bp enhanced green fluorescent protein (EGFP) gene was amplified from the pCT74 vector via PCR. In addition, a 1,086-bp PCR product containing the full-length AfLaeA coding region was amplified from A. flagrans cDNA, and a nourseothricin resistance cassette (Nrsr) was amplified from the pCfB3052 vector. All fragments were sequentially assembled into the pUC19 vector (digested with EcoRI and NdeI) by using the ClonExpress Ultra one-step cloning kit (Vazyme).
For complementation, the experiment was completed using the native promoter of the AfLaeA gene to complement the AfLaeA gene in the mutant strain. In brief, a 1,086-bp PCR product containing the AfLaeA coding region and 2.0-kb upstream and 1.0-kb downstream regions was amplified using the specific primers (Table S8). Similarly, the nourseothricin resistance cassette (Nrsr) was amplified from the pCfB3052 vector, and all fragments were sequentially assembled into the pUC19 vector by the same method as used for overexpression.
Protoplast transformation of A. flagrans.
The transformation system was adapted as previously described (38). In brief, 1 × 108 chlamydospores were inoculated into 100 mL of potato dextrose broth (PDB) and incubated at 28°C and 180 rpm for 24 h. Subsequently, the mycelium was harvested and washed with STC buffer (1 M sorbitol, 50 mM CaCl2, 10 mM Tris-HCl), and approximately 1 g of mycelium (wet weight) was collected and suspended in 2 mL MN buffer (0.3 M MgSO4, 0.3 M CaCl2) containing 1 mg/mL lysing enzyme (Sigma), 2 mg/mL Snailase (Solarbio), and 2 mg/mL cellulase (Solarbio), followed by incubation at 28°C and 90 rpm for 4 h. Subsequently, the protoplasts were filtered using six layers of lens wiping paper and precipitated at 3,000 × g for 10 min. The protoplasts were washed with 10 mL of sorbitol-Tris-calcium chloride (STC) buffer twice, and 200 μL of protoplasts (about 2 × 108) was left at the bottom of the tube, after which 6 to 10 μg of DNA fragment was mixed into 100 μL of protoplasts and incubated on ice for 30 min. One milliliter of PTC (10 mM Tris-HCl [pH 7.5], 50 mM CaCl2, 50% [wt/vol] polyethylene glycol [PEG] 3350) was added and incubated at 28°C for 40 min. Finally, PDSSA medium (24 g/L potato dextrose broth, 0.6 M sucrose, 0.3 g/L peptone, 0.3 g/L tryptone, 0.3 g/L yeast extract, 8 g/L agar) was added to the transformation samples. After the petri dishes were incubated at 28°C for 2 days, the PDA medium containing 100 μg/mL hygromycin B or 50 μg/mL nourseothricin was added to the petri dishes to screen for the positive strains.
Comparison of mycelial growth and analysis of stress tolerance.
WT and ΔAfLaeA strains (6 mm in diameter) were cultivated on PDA, TG (1% tryptone, 1% glucose, 1.5% agar), and TYGA (1% tryptone, 0.5% yeast extract, 1% glucose, 0.5% molasses, 1.5% agar) media for 5 days at 28°C, and the colony diameters were measured daily. For stress tolerance analysis, WT and ΔAfLaeA strains (6 mm in diameter) were cultivated on PDA medium containing chemical stress reagents at different concentration for 5 days at 28°C. In brief, osmotic pressure stress reagent NaCl (0.1, 0.2 and 0.3 M) and cell wall stress reagent SDS (0.01% to 0.03%) were added to the PDA medium. All the experiments were performed at least three times, and the relative growth inhibition (RGI) was calculated using the diameter of each colony.
Trap formation and pathogenicity assays.
For traps, after WT and ΔAfLaeA strains (6 mm in diameter) were cultivated on water agar (WA) medium (2% agar–water) for 3 days at 28°C, approximately 400 to 600 second larval stage (L2) of C. elegans was added to the petri dishes (6 cm in diameter) to induce trap formation, and after the addition of nematodes for 3 h, the trap formation and number of captured nematodes were observed until 96 h. The shape and number of traps were recorded using a microscope and SEM. For extracellular protease activity, the WT, ΔAfLaeA, and OEAfLaeA strains (6 mm in diameter) were inoculated into 100 mL of LMZ medium [gelatin, 20 g/L; peptone, 8 g/L; yeast extract, 1 g/L; (NH4)2SO4, 0.5g/L; MgSO4, 0.5g/L; FeSO4, 0.01 g/L] for 7 days at 180 rpm and 28°C. Ten percent skim milk was added to the WA medium to generate the skim milk dishes, and a puncher was used to prepare the 6-mm hole. Then, 200 μL of fermentation broth of the WT, ΔAfLaeA, and OEAfLaeA strains and TYGA liquid medium were added to the holes. Subsequently, the petri dishes were store at 37°C for 24 h, and the hydrolysis circles were observed under a microscope (Carl Zeiss, Germany).
Cell wall, lipid, glycogen, and nucleic acid staining.
As previously described (32), calcofluor white (Sigma) (20 μg/mL) and 4′,6-diamidino-2-phenylindole (DAPI) (20 μg/mL) were used to stain the cell wall and nuclei, respectively. Glycogen of A. flagrans hyphae and chlamydospores was stained with Lugol’s iodine (Sigma) for 3 min. In addition, LDs were stained with 30 μL of 10 μg/mL BODIPY staining solution (Sigma) for 30 min. Subsequently, the samples were washed twice with phosphate-buffered saline (PBS). The pictures were observed under a fluorescence microscope (Carl Zeiss, Germany).
Real-time fluorescent quantitative PCR assay.
Real-time fluorescent quantitative PCR was performed using AceQ qPCR SYBR green master mix (Vazyme) on a Roche LightCycler 480 system (Roche Applied Science) with the gene-specific primer pairs (Table S9). The amplification conditions were 94°C for 5 s and 60°C for 30 s for 40 cycles. The glyceraldehyde-3-phosphate dehydrogenase (AfGpd) gene was used as an internal control, and the 2−ΔΔCT method was used to calculate the relative transcription level. All assays were repeated at least three times.
Y2H assay.
In order to confirm the interaction between AfLaeA and the predicted proteins, the Y2H assay was used in this study. According to the manufacturer’s instructions (Clontech), the cDNA fragments of AfLaeA were inserted into the pGBKT7 plasmid to generate bait vectors (AfLaeA-BD), whereas the AfVosA, AfVeA, AfVelB, AfVelC, AfPacC, AfFphA, AfBrlA, and AfKapA cDNA fragments were inserted into the pGADT7 plasmid to generate the prey vectors AfVosA-AD, AfVeA-AD, AfVelB-AD, AfVelC-AD, AfPacC-AD, AfFphA-AD, AfBrlA-AD, and AfKapA-AD, respectively. These plasmids were transformed into AH109 yeast cells using a yeast transformation kit (Coolaber), plating on synthetic dropout medium lacking Trp and Leu (SD/–Trp/–Leu), and incubation for 3 days at 30°C. BD-53 and AD-T vector pairs were transformed into AH109 yeast cells to generate positive controls, whereas the empty BD and AD vectors were used as negative controls. The positive yeast cells were selected on SD/–Trp/–Leu medium and transferred to SD/–Trp/–Leu/–His/–Ade medium with serial dilutions (1, 1/20, and 1/100) to determine protein interactions between different pairs.
LC-MS assays.
The secondary metabolites of WT and ΔAfLaeA strains were analyzed by an LC-MS assay in this study. WT and ΔAfLaeA strains were cultured using TYGA liquid medium for 7 days, respectively. The fermentation broths were collected by filtration using a vacuum filter pump. Then, the fermentation broths were extracted three times by mixing them with the same volume of ethyl acetate. The extracts were evaporated under vacuum and dissolved in chromatography-grade methanol (SK, Korea). Finally, the samples were filtered through a 0.22-μm filter and subjected to LC-MS (Thermo Scientific Ultimate 3000; Thermo Fisher Scientific, USA). The metabolic profiles of the WT and ΔAfLaeA strains were compared using Thermo Xcalibur software (Thermo Fisher Scientific). Untargeted metabolomics was performed using Compounds Discoverer 3.0 software (Thermo Fisher Scientific).
SEM and TEM assays.
In this study, SEM was used to observe the differences in morphology, chlamydospores, and traps between WT and ΔAfLaeA strains, while TEM was used to observe the differences in the mycelial interior structure (LDs and EDs) at the hyphal stage, the chlamydospore-producing stage, and the chlamydospore, respectively. In order to analyze the differences in morphology, WT and ΔAfLaeA strains were cultivated on PDA medium with the dialysis membrane (9 cm in diameter) on the surface for 4 days at 28°C. To observe the capacity of chlamydospore production, WT and ΔAfLaeA strains were cultivated on WA medium (2% agar–water) for 8 days at 28°C. For the SEM assay, the samples were fixated with 4% glutaraldehyde for 20 min and dehydrated using an ethanol gradient (70%, 80%, 90%, and 100%). Then, isoamyl acetate (Sigma) was used to treat the sample for 10 min. After drying with liquid carbon dioxide, the samples were observed by SEM. For the TEM assay, the samples were fixed with 2.5% glutaraldehyde and stored at 4°C for at least 12 h. Afterward, the samples were observed under a TEM.
Transcriptome sequencing and analysis.
To compare the effects of the AfLaeA gene on the expression of related genes in A. flagrans, WT and ΔAfLaeA strains were cultured using TYGA liquid medium at 28°C for 3 and 7 days, respectively. Then, the samples were collected and frozen in liquid nitrogen and stored at −80°C. Sequencing of mycelial samples was performed by Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China), and the data were analyzed by the Majorbio Cloud Platform (https:www.majorbio.com). DESeq2 software was used to analyze the DEGs, and the analysis of the conditions of the DEGs included up/down differential multiples of >2 and an adjusted P value of <0.05.
Statistical analyses.
In this study, all experiments were repeated at least three times, and statistical analyses were performed using GraphPad Prism v8.3. The data are expressed as mean values ± standard deviations. One-way analysis of variance, followed by Tukey’s test, was performed to determine significance.
Data availability.
All data generated or analyzed during this study are included in the published paper and the associated supplemental files. The genes numbered “DFL_” and “EVM” correspond to the Duddingtonia (Arthrobotrys) flagrans genome of BioProject accession no. PRJNA494930 and PRJNA917252, respectively. The RNA-seq data presented here are associated with NCBI BioProject PRJNA970849 and BioSample SAMN34997886. The whole-genome shotgun project was deposited in GenBank under BioProject accession no. PRJNA917252 and BioSample accession no. SAMN32532839.
ACKNOWLEDGMENTS
Funding for this study was provided by the Science and Technology special project from Southwest United Graduate School of Yunnan province (grant no. 202302CC4040021), the National Natural Science Foundation of China (grant no. 32160012 and 31860015), and projects from the Department of Science and Technology of Yunnan Province (grant no. 202001BB050061, 202201BC070004, and 202102AA100013).
We are grateful to the Microbial Library of the Germplasm Bank of Wild Species from Southwest China for preserving and providing experimental strains and Guo Yingqi (Kunming Institute of Zoology, Chinese Academy of Sciences) for her help in taking and analyzing TEM images.
We declare we have no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Guo-Hong Li, Email: ligh@ynu.edu.cn.
Mostafa Zamanian, University of Wisconsin-Madison.
REFERENCES
- 1.Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon NJ, Keller NP, Yu JH, Braus GH. 2008. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 2.Bok JW, Keller NP. 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell 3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X, Hu Y, Liu G, Liu M, Li Z, Zhao J, Song X, Zhong Y, Qu Y, Wang L, Qin Y. 2022. The complex Tup1-Cyc8 bridges transcription factor ClrB and putative histone methyltransferase LaeA to activate the expression of cellulolytic genes. Mol Microbiol 117:1002–1022. doi: 10.1111/mmi.14885. [DOI] [PubMed] [Google Scholar]
- 4.Kadooka C, Nakamura E, Mori K, Okutsu K, Yoshizaki Y, Takamine K, Goto M, Tamaki H, Futagami T. 2020. LaeA controls citric acid production through regulation of the citrate exporter-encoding cexA gene in Aspergillus luchuensis mut. kawachii. Appl Environ Microbiol 86:e01950-19. doi: 10.1128/AEM.01950-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Zhang X, Zhang K, Zhang X, Zhu T, Che Q, Zhang G, Li D. 2020. Overexpression of global regulator PbrlaeA leads to the discovery of new polyketide in fungus Penicillium brocae HDN-12-143. Front Chem 8:270. doi: 10.3389/fchem.2020.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martín JF. 2017. Key role of LaeA and velvet complex proteins on expression of β-lactam and PR-toxin genes in Penicillium chrysogenum: cross-talk regulation of secondary metabolite pathways. J Ind Microbiol Biotechnol 44:525–535. doi: 10.1007/s10295-016-1830-y. [DOI] [PubMed] [Google Scholar]
- 7.Ding Z, Wang X, Kong FD, Huang HM, Zhao YN, Liu M, Wang ZP, Han J. 2020. Overexpression of global regulator Talae1 leads to the discovery of new antifungal polyketides from endophytic fungus Trichoderma afroharzianum. Front Microbiol 11:622785. doi: 10.3389/fmicb.2020.622785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fekete E, Karaffa L, Karimi Aghcheh R, Németh Z, Fekete E, Orosz A, Paholcsek M, Stágel A, Kubicek CP. 2014. The transcriptome of lae1 mutants of Trichoderma reesei cultivated at constant growth rates reveals new targets of LAE1 function. BMC Genomics 15:447. doi: 10.1186/1471-2164-15-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahnama M, Maclean P, Fleetwood DJ, Johnson RD. 2019. Comparative transcriptomics analysis of compatible wild type and incompatible ΔlaeA mutant strains of Epichloë festucae in association with perennial ryegrass. Data Brief 24:103843. doi: 10.1016/j.dib.2019.103843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chettri P, Bradshaw RE. 2016. LaeA negatively regulates dothistromin production in the pine needle pathogen Dothistroma septosporum. Fungal Genet Biol 97:24–32. doi: 10.1016/j.fgb.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Estiarte N, Lawrence CB, Sanchis V, Ramos AJ, Crespo-Sempere A. 2016. LaeA and VeA are involved in growth morphology, asexual development, and mycotoxin production in Alternaria alternata. Int J Food Microbiol 238:153–164. doi: 10.1016/j.ijfoodmicro.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 12.López-Díaz C, Rahjoo V, Sulyok M, Ghionna V, Martín-Vicente A, Capilla J, Di Pietro A, López-Berges MS. 2018. Fusaric acid contributes to virulence of Fusarium oxysporum on plant and mammalian hosts. Mol Plant Pathol 19:440–453. doi: 10.1111/mpp.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butchko RA, Brown DW, Busman M, Tudzynski B, Wiemann P. 2012. Lae1 regulates expression of multiple secondary metabolite gene clusters in Fusarium verticillioides. Fungal Genet Biol 49:602–612. doi: 10.1016/j.fgb.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Saha P, Ghosh S, Roy-Barman S. 2020. MoLAEA regulates secondary metabolism in Magnaporthe oryzae. mSphere 5:e00936-19. doi: 10.1128/mSphere.00936-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Y, Yin Z, Wu Y, Xu L, Du H, Wang N, Huang L. 2020. LaeA controls virulence and secondary metabolism in apple canker pathogen Valsa mali. Front Microbiol 11:581203. doi: 10.3389/fmicb.2020.581203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takao K, Akagi Y, Tsuge T, Harimoto Y, Yamamoto M, Kodama M. 2016. The global regulator LaeA controls biosynthesis of host-specific toxins, pathogenicity and development of Alternaria alternata pathotypes. J Gen Plant Pathol 82:121–131. doi: 10.1007/s10327-016-0656-9. [DOI] [Google Scholar]
- 17.Wu D, Oide S, Zhang N, Choi MY, Turgeon BG. 2012. ChLae1 and ChVel1 regulate T-toxin production, virulence, oxidative stress response, and development of the maize pathogen Cochliobolus heterostrophus. PLoS Pathog 8:e1002542. doi: 10.1371/journal.ppat.1002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar D, Barad S, Chen Y, Luo X, Tannous J, Dubey A, Glam Matana N, Tian S, Li B, Keller N, Prusky D. 2017. LaeA regulation of secondary metabolism modulates virulence in Penicillium expansum and is mediated by sucrose. Mol Plant Pathol 18:1150–1163. doi: 10.1111/mpp.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarikaya Bayram O, Bayram O, Valerius O, Park HS, Irniger S, Gerke J, Ni M, Han KH, Yu JH, Braus GH. 2010. LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet 6:e1001226. doi: 10.1371/journal.pgen.1001226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayram O, Braus GH. 2012. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev 36:1–24. doi: 10.1111/j.1574-6976.2011.00285.x. [DOI] [PubMed] [Google Scholar]
- 21.Liu Q, Cai L, Shao Y, Zhou Y, Li M, Wang X, Chen F. 2016. Inactivation of the global regulator LaeA in Monascus ruber results in a species-dependent response in sporulation and secondary metabolism. Fungal Biol 120:297–305. doi: 10.1016/j.funbio.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Lan N, Yue Q, An Z, Bills GF. 2020. Apc.LaeA and Apc.VeA of the velvet complex govern secondary metabolism and morphological development in the echinocandin-producing fungus Aspergillus pachycristatus. J Ind Microbiol Biotechnol 47:155–168. doi: 10.1007/s10295-019-02250-x. [DOI] [PubMed] [Google Scholar]
- 23.Crespo-Sempere A, Marín S, Sanchis V, Ramos AJ. 2013. VeA and LaeA transcriptional factors regulate ochratoxin A biosynthesis in Aspergillus carbonarius. Int J Food Microbiol 166:479–486. doi: 10.1016/j.ijfoodmicro.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 24.Karimi Aghcheh R, Németh Z, Atanasova L, Fekete E, Paholcsek M, Sándor E, Aquino B, Druzhinina IS, Karaffa L, Kubicek CP. 2014. The VELVET A orthologue VEL1 of Trichoderma reesei regulates fungal development and is essential for cellulase gene expression. PLoS One 9:e112799. doi: 10.1371/journal.pone.0112799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi JC, Shi WL, Zhou YR, Chen XL, Zhang YZ, Zhang X, Zhang WX, Song XY. 2020. The putative methyltransferase TlLAE1 is involved in the regulation of peptaibols production in the biocontrol fungus Trichoderma longibrachiatum SMF2. Front Microbiol 11:1267. doi: 10.3389/fmicb.2020.01267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu C, Wang Y, Hu X, Lei M, Wang M, Zeng J, Li H, Liu Z, Zhou T, Yu D. 2020. Involvement of LaeA in the regulation of conidia production and stress responses in Penicillium digitatum. J Basic Microbiol 60:82–88. doi: 10.1002/jobm.201900367. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Zhu Y, Bao L, Gao L, Yao G, Li Y, Yang Z, Li Z, Zhong Y, Li F, Yin H, Qu Y, Qin Y. 2016. Putative methyltransferase LaeA and transcription factor CreA are necessary for proper asexual development and controlling secondary metabolic gene cluster expression. Fungal Genet Biol 94:32–46. doi: 10.1016/j.fgb.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Kosalková K, García-Estrada C, Ullán RV, Godio RP, Feltrer R, Teijeira F, Mauriz E, Martín JF. 2009. The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine C synthesis in Penicillium chrysogenum. Biochimie 91:214–225. doi: 10.1016/j.biochi.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Cheng M, Zhao S, Lin C, Song J, Yang Q. 2021. Requirement of LaeA for sporulation, pigmentation and secondary metabolism in Chaetomium globosum. Fungal Biol 125:305–315. doi: 10.1016/j.funbio.2020.11.008. [DOI] [PubMed] [Google Scholar]
- 30.Luo Q, Li N, Xu JW. 2022. A methyltransferase LaeA regulates ganoderic acid biosynthesis in Ganoderma lingzhi. Front Microbiol 13:1025983. doi: 10.3389/fmicb.2022.1025983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Yang E, An Z, Liu X. 2007. Evolution of nematode-trapping cells of predatory fungi of the Orbiliaceae based on evidence from rRNA-encoding DNA and multiprotein sequences. Proc Natl Acad Sci USA 104:8379–8384. doi: 10.1073/pnas.0702770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Youssar L, Wernet V, Hensel N, Yu X, Hildebrand HG, Schreckenberger B, Kriegler M, Hetzer B, Frankino P, Dillin A, Fischer R. 2019. Intercellular communication is required for trap formation in the nematode-trapping fungus Duddingtonia flagrans. PLoS Genet 15:e1008029. doi: 10.1371/journal.pgen.1008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mei X, Wang X, Li G. 2021. Pathogenicity and volatile nematicidal metabolites from Duddingtonia flagrans against Meloidogyne incognita. Microorganisms 9:2268. doi: 10.3390/microorganisms9112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balbino HM, Gouveia AD, Monteiro TSA, Morgan T, de Freitas LG. 2022. Overview of the nematophagous fungus Duddingtonia flagrans. Biocontrol Sci Technol 32:911–929. doi: 10.1080/09583157.2022.2094891. [DOI] [Google Scholar]
- 35.Wernet V, Fischer R. 2023. Establishment of Arthrobotrys flagrans as biocontrol agent against the root pathogenic nematode Xiphinema index. Environ Microbiol 25:283–293. doi: 10.1111/1462-2920.16282. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C, Zhang H, Zhu Q, Hao S, Chai S, Li Y, Jiao Z, Shi J, Sun B, Wang C. 2020. Overexpression of global regulator LaeA increases secondary metabolite production in Monascus purpureus. Appl Microbiol Biotechnol 104:3049–3060. doi: 10.1007/s00253-020-10379-4. [DOI] [PubMed] [Google Scholar]
- 37.Wernet N, Wernet V, Fischer R. 2021. The small-secreted cysteine-rich protein CyrA is a virulence factor participating in the attack of Caenorhabditis elegans by Duddingtonia flagrans. PLoS Pathog 17:e1010028. doi: 10.1371/journal.ppat.1010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W, Zhao Y, Bai N, Zhang KQ, Yang J. 2022. AMPK is involved in regulating the utilization of carbon sources, conidiation, pathogenicity, and stress response of the nematode-trapping fungus Arthrobotrys oligospora. Microbiol Spectr 10:e0222522. doi: 10.1128/spectrum.02225-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu X, Hu X, Pop M, Wernet N, Kirschhöfer F, Brenner-Weiß G, Keller J, Bunzel M, Fischer R. 2021. Fatal attraction of Caenorhabditis elegans to predatory fungi through 6-methyl-salicylic acid. Nat Commun 12:5462. doi: 10.1038/s41467-021-25535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang L, Li X, Bai N, Yang X, Zhang KQ, Yang J. 2022. Transcriptomic analysis reveals that Rho GTPases regulate trap development and lifestyle transition of the nematode-trapping fungus Arthrobotrys oligospora. Microbiol Spectr 10:e0175921. doi: 10.1128/spectrum.01759-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang KX, Liu QQ, Bai N, Zhu MC, Zhang KQ, Yang JK. 2022. AoSsk1, a response regulator required for mycelial growth and development, stress responses, trap formation, and the secondary metabolism in Arthrobotrys oligospora. J Fungi (Basel) 8:260. doi: 10.3390/jof8030260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeitz MA, Tanveer Z, Openshaw AT, Schmidt M. 2019. Genetic regulators and physiological significance of glycogen storage in Candida albicans. J Fungi (Basel) 5:102. doi: 10.3390/jof5040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bromley M, Johns A, Davies E, Fraczek M, Mabey Gilsenan J, Kurbatova N, Keays M, Kapushesky M, Gut M, Gut I, Denning DW, Bowyer P. 2016. Mitochondrial complex I is a global regulator of secondary metabolism, virulence and azole sensitivity in fungi. PLoS One 11:e0158724. doi: 10.1371/journal.pone.0158724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu QY, Pu XJ, Li GH, Li CQ, Lei HM, Zhang KQ, Zhao PJ. 2022. Identification and mechanism of action of the global secondary metabolism regulator SaraC in Stereum hirsutum. Microbiol Spectr 10:e0262422. doi: 10.1128/spectrum.02624-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei Z, Shu D, Sun Q, Chen DB, Li ZM, Luo D, Yang J, Tan H. 2022. The BcLAE1 is involved in the regulation of ABA biosynthesis in Botrytis cinerea TB-31. Front Microbiol 13:969499. doi: 10.3389/fmicb.2022.969499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wernet V, Wäckerle J, Fischer R. 2022. The STRIPAK component SipC is involved in morphology and cell-fate determination in the nematode-trapping fungus Duddingtonia flagrans. Genetics 220:iyab153. doi: 10.1093/genetics/iyab153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akhberdi O, Zhang Q, Wang D, Wang H, Hao X, Liu Y, Wei D, Zhu X. 2018. Distinct roles of velvet complex in the development, stress tolerance, and secondary metabolism in Pestalotiopsis microspora, a taxol producer. Genes (Basel) 9:164. doi: 10.3390/genes9030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsunematsu Y, Takanishi J, Asai S, Masuya T, Nakazawa T, Watanabe K. 2019. Genomic mushroom hunting decrypts coprinoferrin, a siderophore secondary metabolite vital to fungal cell development. Org Lett 21:7582–7586. doi: 10.1021/acs.orglett.9b02861. [DOI] [PubMed] [Google Scholar]
- 49.Park HS, Ni M, Jeong KC, Kim YH, Yu JH. 2012. The role, interaction and regulation of the velvet regulator VelB in Aspergillus nidulans. PLoS One 7:e45935. doi: 10.1371/journal.pone.0045935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang G, Zheng Y, Ma Y, Yang L, Xie M, Zhou D, Niu X, Zhang KQ, Yang J. 2019. The velvet proteins VosA and VelB play different roles in conidiation, trap formation, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Front Microbiol 10:1917. doi: 10.3389/fmicb.2019.01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin SW, Douglas LM, Konopka JB. 2005. Cell cycle dynamics and quorum sensing in Candida albicans chlamydospores are distinct from budding and hyphal growth. Eukaryot Cell 4:1191–1202. doi: 10.1128/EC.4.7.1191-1202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kües U, Walser PJ, Klaus MJ, Aebi M. 2002. Influence of activated A and B mating-type pathways on developmental processes in the basidiomycete Coprinus cinereus. Mol Genet Genomics 268:262–271. doi: 10.1007/s00438-002-0745-7. [DOI] [PubMed] [Google Scholar]
- 53.Staib P, Morschhäuser J. 2007. Chlamydospore formation in Candida albicans and Candida dubliniensis—an enigmatic developmental programme. Mycoses 50:1–12. doi: 10.1111/j.1439-0507.2006.01308.x. [DOI] [PubMed] [Google Scholar]
- 54.Citiulo F, Moran GP, Coleman DC, Sullivan DJ. 2009. Purification and germination of Candida albicans and Candida dubliniensis chlamydospores cultured in liquid media. FEMS Yeast Res 9:1051–1060. doi: 10.1111/j.1567-1364.2009.00533.x. [DOI] [PubMed] [Google Scholar]
- 55.Bae YS, Knudsen GR. 2000. Cotransformation of Trichoderma harzianum with beta-glucuronidase and green fluorescent protein genes provides a useful tool for monitoring fungal growth and activity in natural soils. Appl Environ Microbiol 66:810–815. doi: 10.1128/AEM.66.2.810-815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ment D, Gindin G, Glazer I, Perl S, Elad D, Samish M. 2010. The effect of temperature and relative humidity on the formation of Metarhizium anisopliae chlamydospores in tick eggs. Fungal Biol 114:49–56. doi: 10.1016/j.mycres.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 57.Kües U. 2000. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev 64:316–353. doi: 10.2307/3758110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Böttcher B, Pöllath C, Staib P, Hube B, Brunke S. 2016. Candida species rewired hyphae developmental programs for chlamydospore formation. Front Microbiol 7:1697. doi: 10.3389/fmicb.2016.01697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hernández-Cervantes A, Znaidi S, van Wijlick L, Denega I, Basso V, Ropars J, Sertour N, Sullivan D, Moran G, Basmaciyan L, Bon F, Dalle F, Bougnoux ME, Boekhout T, Yang Y, Li Z, Bachellier-Bassi S, d'Enfert C. 2020. A conserved regulator controls asexual sporulation in the fungal pathogen Candida albicans. Nat Commun 11:6224. doi: 10.1038/s41467-020-20010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hou YH, Hsu LH, Wang HF, Lai YH, Chen YL. 2020. Calcineurin regulates conidiation, chlamydospore formation and virulence in Fusarium oxysporum f. sp. lycopersici. Front Microbiol 11:539702. doi: 10.3389/fmicb.2020.539702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng X, Wu B, Zhang S, Li M, Jiang X. 2021. Transcriptome dynamics underlying chlamydospore formation in Trichoderma virens GV29-8. Front Microbiol 12:654855. doi: 10.3389/fmicb.2021.654855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li P, Cao X, Zhang L, Lv M, Zhang LH. 2022. PhcA and PhcR regulate ralsolamycin biosynthesis oppositely in Ralstonia solanacearum. Front Plant Sci 13:903310. doi: 10.3389/fpls.2022.903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elramli N, Karahoda B, Sarikaya-Bayram Ö, Frawley D, Ulas M, Oakley CE, Oakley BR, Seiler S, Bayram Ö. 2019. Assembly of a heptameric STRIPAK complex is required for coordination of light-dependent multicellular fungal development with secondary metabolism in Aspergillus nidulans. PLoS Genet 15:e1008053. doi: 10.1371/journal.pgen.1008053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu J, He X, Xu C, Yu M, Song T, Cao H, Pan X, Qi Z, Du Y, Zhang R, Liang D, Liu Y. 2022. Autophagy-related protein UvAtg7 contributes to mycelial growth, virulence, asexual reproduction and cell stress response in rice false smut fungus Ustilaginoidea virens. Fungal Genet Biol 159:103668. doi: 10.1016/j.fgb.2022.103668. [DOI] [PubMed] [Google Scholar]
- 65.Yu J, Yu M, Song T, Cao H, Pan X, Yong M, Qi Z, Du Y, Zhang R, Yin X, Liu Y. 2019. A homeobox transcription factor UvHOX2 regulates chlamydospore formation, conidiogenesis, and pathogenicity in Ustilaginoidea virens. Front Microbiol 10:1071. doi: 10.3389/fmicb.2019.01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu M, Yu J, Cao H, Pan X, Song T, Qi Z, Du Y, Huang S, Liu Y. 2022. The velvet protein UvVEA regulates conidiation and chlamydospore formation in Ustilaginoidea virens. J Fungi (Basel) 8:479. doi: 10.3390/jof8050479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cai YY, Wang JY, Wu XY, Liang S, Zhu XM, Li L, Lu JP, Liu XH, Lin FC. 2022. MoOpy2 is essential for fungal development, pathogenicity, and autophagy in Magnaporthe oryzae. Environ Microbiol 24:1653–1671. doi: 10.1111/1462-2920.15949. [DOI] [PubMed] [Google Scholar]
- 68.Bhutada G, Kavšcek M, Ledesma-Amaro R, Thomas S, Rechberger GN, Nicaud JM, Natter K. 2017. Sugar versus fat: elimination of glycogen storage improves lipid accumulation in Yarrowia lipolytica. FEMS Yeast Res 17:fox020. doi: 10.1093/femsyr/fox020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oda K, Kobayashi A, Ohashi S, Sano M. 2011. Aspergillus oryzae laeA regulates kojic acid synthesis genes. Biosci Biotechnol Biochem 75:1832–1834. doi: 10.1271/bbb.110235. [DOI] [PubMed] [Google Scholar]
- 70.Niu J, Arentshorst M, Nair PD, Dai Z, Baker SE, Frisvad JC, Nielsen KF, Punt PJ, Ram AF. 2015. Identification of a classical mutant in the industrial host Aspergillus niger by systems genetics: LaeA is required for citric acid production and regulates the formation of some secondary metabolites. G3 (Bethesda) 6:193–204. doi: 10.1534/g3.115.024067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hong EJ, Kim NK, Lee D, Kim WG, Lee I. 2015. Overexpression of the laeA gene leads to increased production of cyclopiazonic acid in Aspergillus fumisynnematus. Fungal Biol 119:973–983. doi: 10.1016/j.funbio.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 72.Karimi-Aghcheh R, Bok JW, Phatale PA, Smith KM, Baker SE, Lichius A, Omann M, Zeilinger S, Seiboth B, Rhee C, Keller NP, Freitag M, Kubicek CP. 2013. Functional analyses of Trichoderma reesei LAE1 reveal conserved and contrasting roles of this regulator. G3 (Bethesda) 3:369–378. doi: 10.1534/g3.112.005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He T, Wang YD, Du LQ, Li FR, Hu QF, Cheng GG, Wang WG. 2020. Overexpression of global regulator LaeA induced secondary metabolite production in Aspergillus versicolor 0312. Rec Nat Prod 14:387–394. doi: 10.25135/rnp.183.20.03.1593. [DOI] [Google Scholar]
- 74.Yu J, Han H, Zhang X, Ma C, Sun C, Che Q, Gu Q, Zhu T, Zhang G, Li D. 2019. Discovery of two new sorbicillinoids by overexpression of the global regulator LaeA in a marine-derived fungus Penicillium dipodomyis YJ-11. Mar Drugs 17:446. doi: 10.3390/md17080446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Müller N, Leroch M, Schumacher J, Zimmer D, Könnel A, Klug K, Leisen T, Scheuring D, Sommer F, Mühlhaus T, Schroda M, Hahn M. 2018. Investigations on VELVET regulatory mutants confirm the role of host tissue acidification and secretion of proteins in the pathogenesis of Botrytis cinerea. New Phytol 219:1062–1074. doi: 10.1111/nph.15221. [DOI] [PubMed] [Google Scholar]
- 76.Qin Y, Ortiz-Urquiza A, Keyhani NO. 2014. A putative methyltransferase, mtrA, contributes to development, spore viability, protein secretion and virulence in the entomopathogenic fungus Beauveria bassiana. Microbiology (Reading) 160:2526–2537. doi: 10.1099/mic.0.078469-0. [DOI] [PubMed] [Google Scholar]
- 77.Lv Y, Lv A, Zhai H, Zhang S, Li L, Cai J, Hu Y. 2018. Insight into the global regulation of laeA in Aspergillus flavus based on proteomic profiling. Int J Food Microbiol 284:11–21. doi: 10.1016/j.ijfoodmicro.2018.06.024. [DOI] [PubMed] [Google Scholar]
- 78.Barrios-González J, Pérez-Sánchez A, Bibián ME. 2020. New knowledge about the biosynthesis of lovastatin and its production by fermentation of Aspergillus terreus. Appl Microbiol Biotechnol 104:8979–8998. doi: 10.1007/s00253-020-10871-x. [DOI] [PubMed] [Google Scholar]
- 79.Strauss J, Reyes-Dominguez Y. 2011. Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet Biol 48:62–69. doi: 10.1016/j.fgb.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.00186-23-s0001.pdf, PDF file, 1.7 MB (1.7MB, pdf)
Data Availability Statement
All data generated or analyzed during this study are included in the published paper and the associated supplemental files. The genes numbered “DFL_” and “EVM” correspond to the Duddingtonia (Arthrobotrys) flagrans genome of BioProject accession no. PRJNA494930 and PRJNA917252, respectively. The RNA-seq data presented here are associated with NCBI BioProject PRJNA970849 and BioSample SAMN34997886. The whole-genome shotgun project was deposited in GenBank under BioProject accession no. PRJNA917252 and BioSample accession no. SAMN32532839.