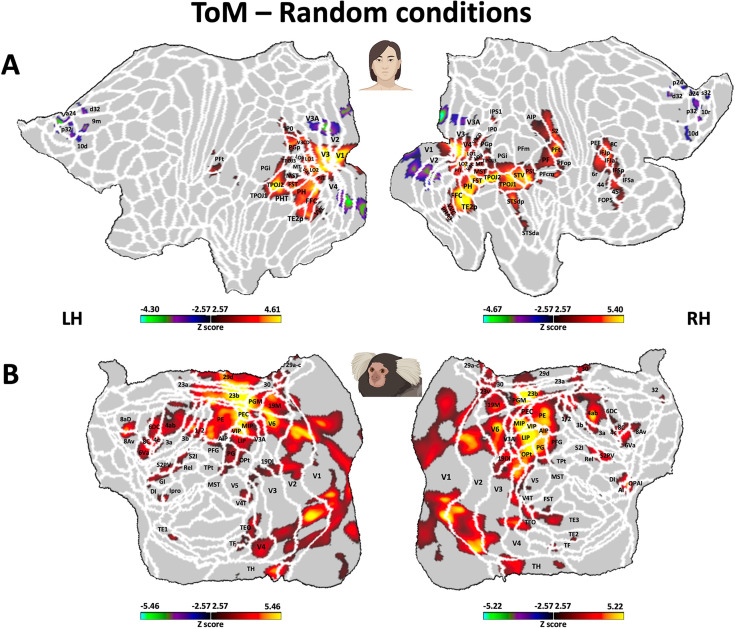
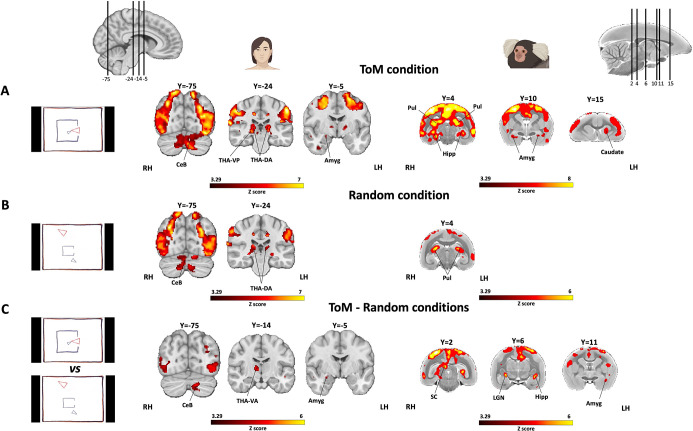
Figure 5. Brain network involved during processing of ToM compared to Random Frith-Happé’s animations in both humans (A) and marmosets (B).
Group functional maps showing significant greater activations for ToM animations compared to Random animations. (A) Group map obtained from 10 human subjects displayed on the left and right human cortical flat maps. The white line delineates the regions based on the recent multi-modal cortical parcellation atlas (Glasser et al., 2016). (B) Group map obtained from 6 marmosets displayed on the left and right marmoset cortical flat maps. The white line delineates the regions based on the Paxinos parcellation of the NIH marmoset brain atlas (Liu et al., 2018). The brain areas reported in A and B have activation threshold corresponding to z-scores >2.57 (yellow/red scale) or z-scores <–2.57 (purple/green scale) (AFNI’s 3dttest++, cluster-forming threshold of p<0.01 uncorrected and then FWE-corrected α=0.05 at cluster-level from 10,000 Monte-Carlo simulations).