Figure 2.
Extra crossovers in Fancm are consistent with the characteristics of class II crossovers
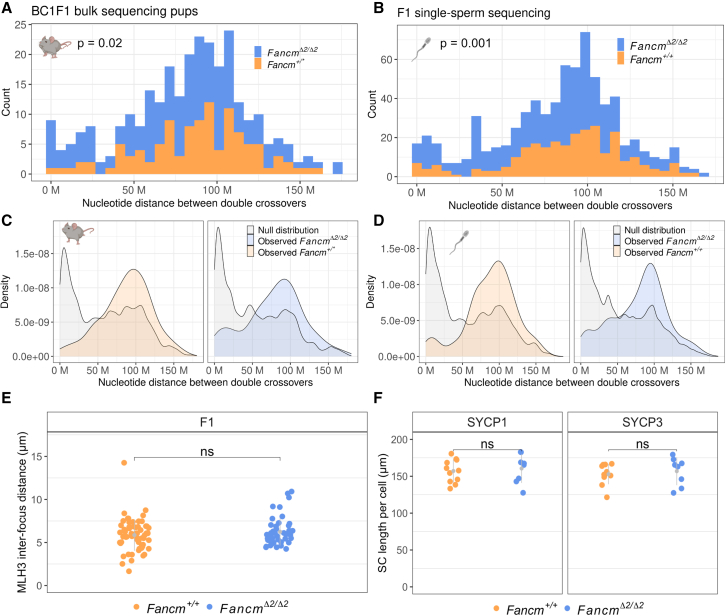
Interference is detected in Fancm-deficient male mice, but extra crossovers fit with a more random distribution and are not labeled by MLH1/3.
(A and B) Intercrossover distances were quantified in the wild type and the mutant from the BC1F1-derived bulk sequencing and sperm sequencing data by including only chromosomes with exactly two crossovers. Printed p values are from Wilcoxon rank-sum test. (A) n = (175 FancmΔ2/Δ2 and 109 Fancm+/∗ double crossovers per group. Fancm+/∗ indicates pooling of Fancm+/+ and Fancm+/Δ2 data, as their crossover rates were shown not to be significantly different). (B) n = (453 FancmΔ2/Δ2 and 276 Fancm+/+).
(C and D) Crossover interference is observed in the wild type and the mutant, as the observed intercrossover distributions are more evenly spaced than an expected random distribution generated via permuting genotype group labels (B = 1,000).
(E) Cytological measurement of interference used MLH3 interfocus distances in both the wild type and the mutant. n = 2 F1 (FVB/N × C57BL/6J) mice per genotype. Total cells analyzed: 62 F1.Fancm+/+ and 47 F1.FancmΔ2/Δ2. Gray bars indicate mean ± SD.
(F) Synaptonemal complex length was assessed by measuring SYCP1 and SYCP3 length in micrometers per nucleus in pachytene spermatocytes. Total cells analyzed: 16 F1.Fancm+/+ and 22 F1.FancmΔ2/Δ2. Gray bars indicate mean ± SD.