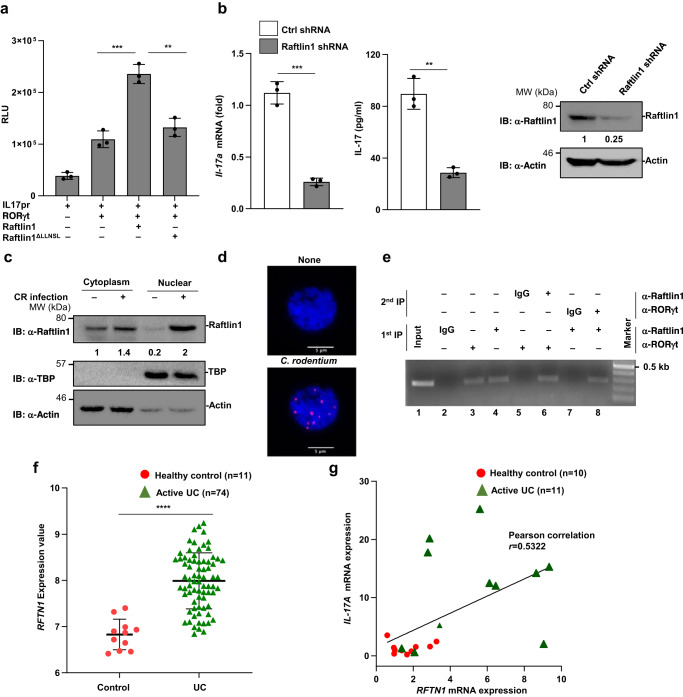
Fig. 2. Raftlin1 regulates IL-17 expression, and the level of Raftlin1 expression correlates with IL-17 expression in UC patients.
a Luciferase assay of lysates of Jurkat T cells transfected with various plasmid combinations (below plot) of the IL-17 promoter-driven luciferase plasmid (pGL4-IL17pr) and the plasmid encoding Flag-RORγt along with wild-type Myc-Raftlin1 or Myc-Raftlin1ΔLLNSL; results are presented in relative luciferase unit (RLU); n = 3 biological replicates, p = 0.0009, p = 0.002. b Real-time PCR and ELISA were performed to check the expression of IL-17 in Raftlin1 knockdown lysates of CD4+ T cells isolated from colonic lamina propria of C. rodentium infected WT mice; n = 3 biological replicates, p = 0.0002 (Il-17a), p = 0.001 (IL-17a). Immunoblot showing knockdown of Raftlin1. c Immunoblot analysis for Raftlin1 in the cytoplasmic and nuclear fraction of CD4+ T cells isolated from colonic lamina propria. d Representative images from proximity ligation assay (PLA) showing the interaction between RORγt and Raftlin1. The image is representative of three independent experiments. Scale bars, 5 μm. e Binding of RORγt-Raftlin1 to the IL-17 promoter sites was assessed by ChIP, and re-ChIP analysis using a DNA-protein complex using a specific antibody. f GEO dataset (GSE59071) was analyzed (n = 11 healthy control and n = 74 active UC patients samples), and the normalized expression value of RFTN1 is as shown above, p = 2 × 10−9. g Expression of IL-17A mRNA and RFTN1 mRNA was analyzed by real-time PCR in active UC and control samples (n = 10 healthy control and n = 11 active UC patients samples). Relative IL-17A mRNA was plotted against relative RFTN1 mRNA; p = 0.0130, Pearson correlation coefficient r = 0.5322. Data in (a–c, e) are from one experiment representing three independent experiments with similar results. Statistical significance was determined by unpaired Student’s t test (two-tailed) in (a, b, f, g) with p < 0.05 considered statistically significant. **p < 0.01; ***p < 0.001; ****p < 0.0001, error bars are mean ± SD. The statistics (g) were measured by the Pearson correlation coefficient. Source data are provided as a Source data file.