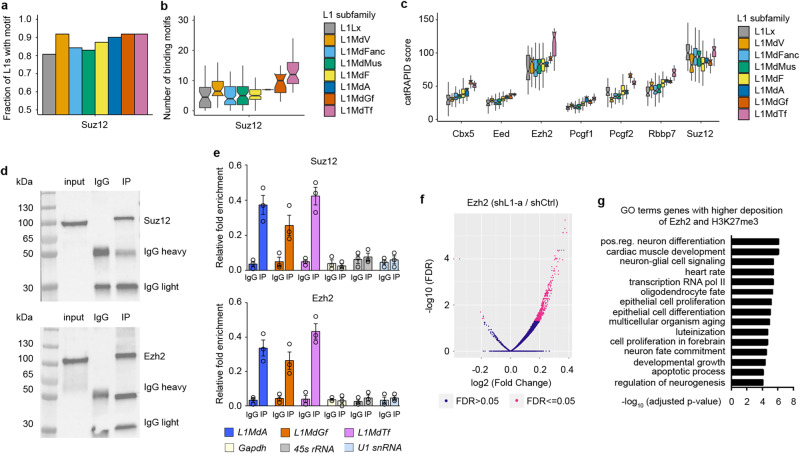
Fig. 4. L1 RNAs bind PRC2 subunits Ezh2 and Suz12 and influences the binding of Ezh2.
a, b Boxplots showing the fraction of L1 sequences per evolutionary time-point hosting a Suz12 RNA binding motif (a) and the number of motifs per L1 detected by catRAPID (b). c Boxplots of catRAPID score for 7 Polycomb proteins along evolutionary time-points. The boxes show the interquartile range (IQR), the central line represents the median, the whiskers add 1.5 times the IQR to the 75 percentile (box upper limit) and subtract 1.5 times the IQR from the 25 percentile (box lower limit). For (a, b and c), the number of different L1 elements for each time-point is the following: L1Lx n = 78, L1MdV n = 26, L1MdFanc n = 38, L1MdMus n = 17, L1MdF n = 205, L1MdA n = 47, L1MdGf n = 47, L1MdTf n = 57. d, e RNA immunoprecipitation (IP) for Suz12 and Ezh2 with chromatinic RNA fraction of 21 div cells. Western blot showing Suz12 and Ezh2 enrichment after IP (d) and RNA levels by RT-qPCR for L1 subfamilies RNAs, Gapdh, 45 s rRNA and U1 snRNA in Suz12 or Ezh2 IP. RNA levels are relative to input control. n = 3 independent biological samples. Data are mean ± s.e.m. f Volcano plots representing differentially enriched ChIP-seq peaks in shL1-a cortical cells for Ezh2. X-axis shows the log2 (Fold Change). The Y-axis shows the -log10 (FDR). n = 2 independent biological replicates. g Top GO terms under the biological process category for genes contained in peaks/islands characterized by a significantly higher deposition of Ezh2 and H3K27me3. p values were determined by gprofiler2 using a default hypergeometric test and correction for multiple testing has been performed by the g:SCS algorithm. Source data are provided as a Source Data file.