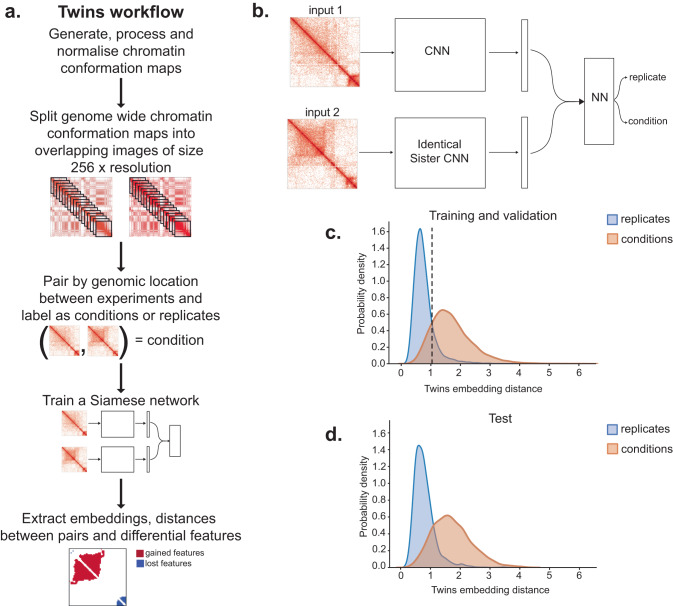
Fig. 1. A replicate-based method for deep learning from chromatin conformation contact maps.
a Overview of the Twins workflow. b Illustration of the Siamese network architecture for chromatin conformation maps. c Twins embedding distance distributions for replicates (blue) and conditions (orange, mature versus immature thymocytes) for training (chromosomes 1, 3–17, 19) and validation (chromosome 18). d As in (c) but on test set (chromosome 2).