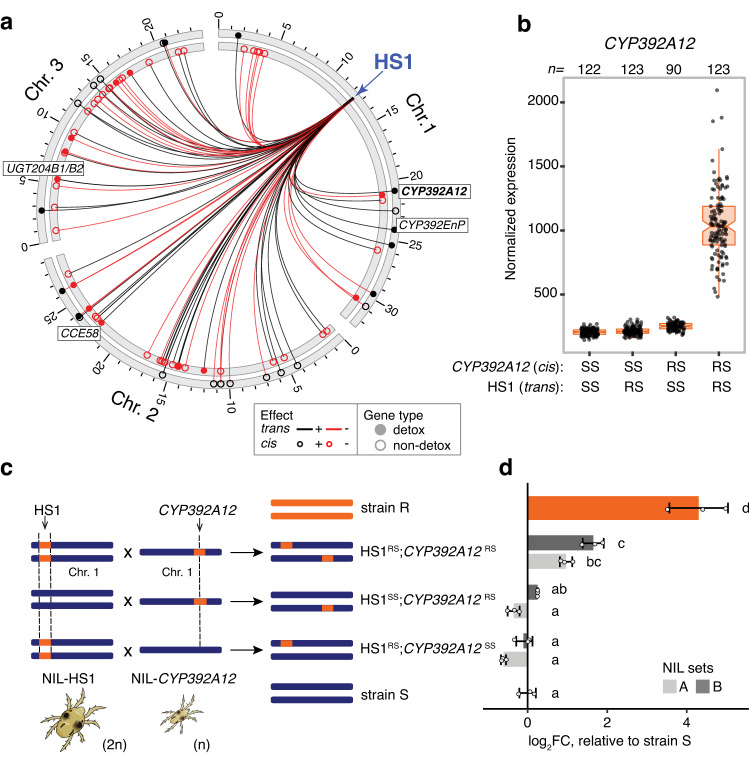
Fig. 4. An epistatic interaction between CYP392A12 trans and cis eQTLs.
a Genes trans regulated by HS1 that also have cis eQTLs are displayed in a circle plot (the lines originating from HS1, blue arrow, link HS1 to the genomic locations of the respective genes). Select detoxification genes (filled circles, legend, bottom right) with large HS1 trans-driven fold changes (CYP392A12, UGT204B1, UGT204B2, CCE58, and CYP392EnP, see Fig. 3c) are labeled, with CYP392A12 in bold text. The direction of effects of trans eQTLs at HS1, as well as cis eQTLs at target genes, is as indicated in the legend; “+” indicates upregulation (black lines for trans, and outer circle for cis) by the RS genotype as compared to the SS genotype (otherwise, “-”, red lines for trans, and inner circle for cis). b A boxplot showing DESeq2 normalized expression of CYP392A12 across the 458 F3 families conditioned on the four possible genotypes at HS1 (reflecting a trans effect) and at CYP392A12 (reflecting a cis effect; sample sizes, n, are given at top). For the boxplot, the box indentations are median values, the boxes extend from the first to third quartiles, and the whiskers extend to 1.5 times the respective interquartile ranges. All data points are displayed. c Schematic showing crosses between the B-NIL-HS1 strain females (2n) and males (n) from the two sets ("A" and "B") of the CYP392A12 NILs. RNA was collected from F1 females and from the parent R and S strains (color coded as orange and blue, respectively). d Displayed are log2 fold change (log2FC) values for CYP392A12 in F1 females from the crosses shown in panel c, as well as strain R, in comparison to that of the S strain (one-way analysis of variance, F7,16 = 82.67, p = 2.29 × 10-11; samples with different letters have significant differences as assessed with pairwise two-sided unpaired t-tests, adj-p < 0.05; p values were corrected for multiple tests with the Bonferroni method). For the bar plot, means, error bars of ±1 standard deviation, and all data points are shown (n = 3 biologically independent replicates, with each biological replicate based on two technical replicates).