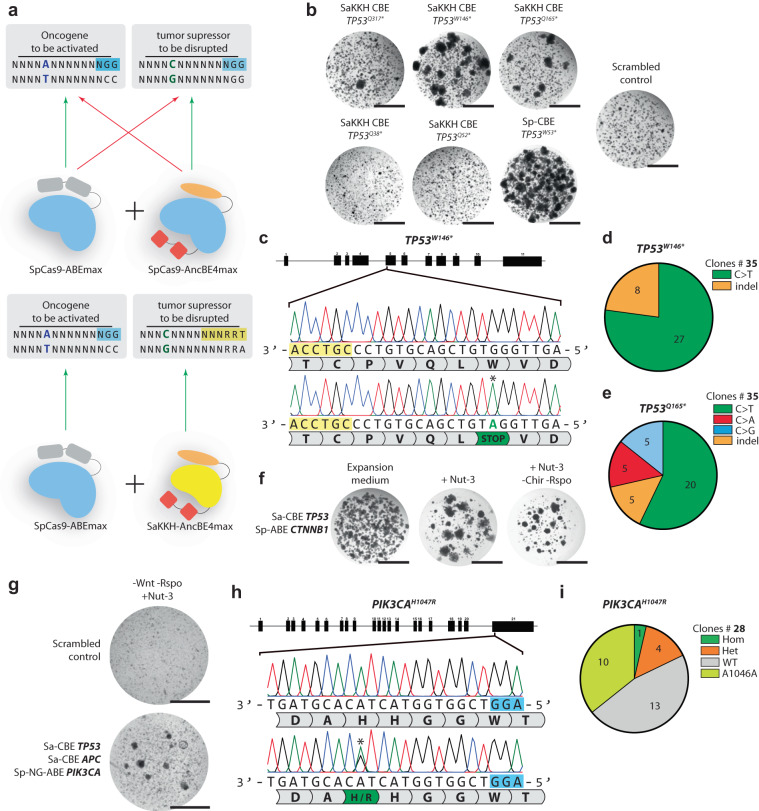
Fig. 5. Simultaneous C > T and A > G engineering by Cas9 homolog multiplexing.
a Schematic overview of the problem arising when CBE and ABE is applied using the same cas9-homolog. This can be circumvented by using Sa-CBE together with Sp-ABE. b Brightfield images showing hepatocyte organoid outgrowth upon Nutlin-3 selection on 3 SaKKH-sgRNA’s compared to a scrambled control sgRNA and our optimal TP53W53* sgRNA in combination with SpCas9-CBE. Scale bars are 2000 µm. c Sanger sequencing of hepatocyte organoids surviving Nutlin-3 selection upon transfection of SaKKH-CBE and a TP53W146* sgRNA. Edited residue is highlighted with an asterisk and SaKKH PAM is shown in yellow. d Quantification of editing observed in Nutlin-3 resistant clones highlights increased indel induction by SaKKH-CBE. Clones with homozygous C > T mutations are shown in green and clones with indels on one of their alleles are shown in orange. e Quantification of editing observed in Nutlin-3 resistant clones highlights increased amount of unintended edits upon SaKKH-CBE editing. Pie-chart shows clones with homozygous C > T (green) mutations and clones harboring a C > A (red), C > G (blue), or indel (orange) on at least one of their alleles. f Brightfield images of organoids upon Cas9-homolog multiplexing using SpCas9-ABE to target CTNNB1 and SaKKHcas9-CBE to target TP53. Organoids are selected for TP53 mutations by addition of Nutlin-3 to the culture media, and selected for CTNNB1 mutations by addition removal of CHIR and Rspondin-1. Scale bars are 2000 µm. g Brightfield images of Cas9-homolog multiplexing. Organoids are transfected with SaKKH-CBE targeting TP53 and APC and SpCas9-NG targeting PIK3CAH1047R. Organoids are selected on medium lacking wnt and Rspondin-1 (rpso) and with the addition of Nutlin-3(Nut-3)Scale bars are 2000 µm. h Sanger sequencing of the PIK3CAH1047R locus showing heterozygous mutation induction. PAM is shown in blue and mutation is highlighted with an asterisk. i Quantification of editing efficiency of the SpCas9-NG PIK3CAH1047R sgRNA by Sanger sequencing. Hom homozygous, Het heterozygous, WT wild type. Source data are provided as a Source data file.