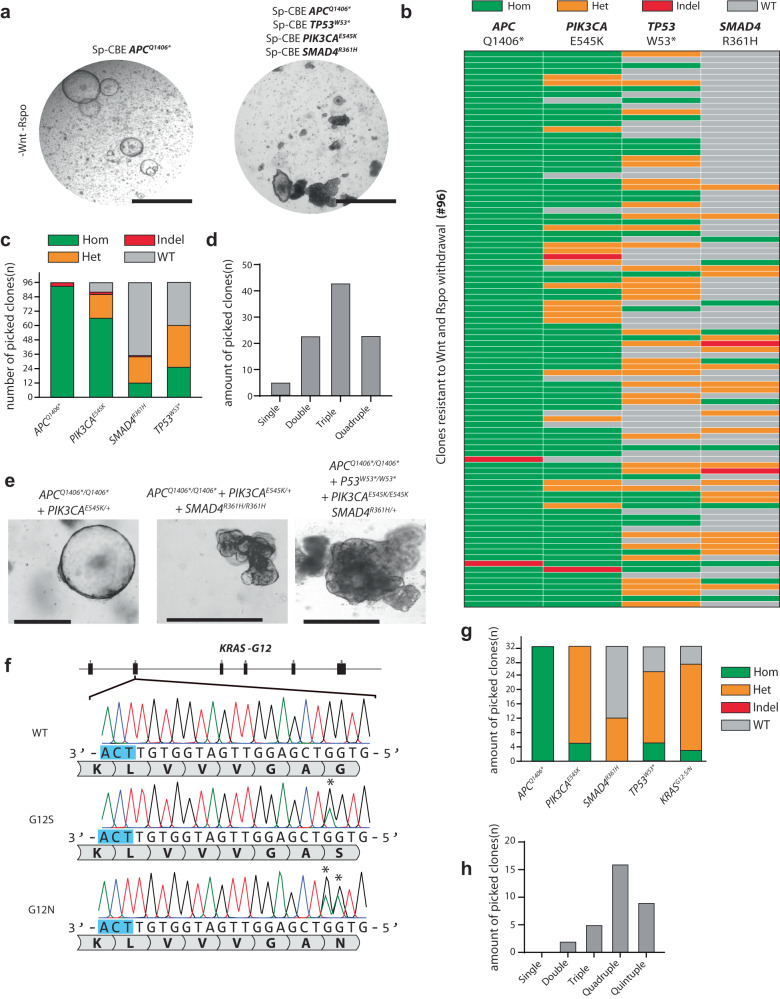
Fig. 6. One-step generation of a mini-biobank containing mutants that recapitulate intermittent steps of colorectal cancer tumorigenesis.
a Brightfield images of intestinal organoids transfected with only SpCas9-CBE and a sgRNA targeting APCQ1406* and organoids transfected with our cocktail of 4 sgRNA’s targeting APC, TP53, SMAD4, and PIK3CA. Scale bars are 2000 µm. b Sanger sequencing of 96 clones surviving Wnt-pathway selection by removal of Rspondin-1 and Wnt-surrogate from culture medium. Starting with a cystic structure (top) to a more dense phenotype (bottom). c Quantification of editing efficiency of our 4 sgRNA by Sanger sequencing. d Quantification of single, double, triple, and quadruple mutants acquired from a single transfection experiment in human intestinal organoids. e Brightfield images highlighting increasingly dense morphology upon introduction of additional mutations on top of APC. Scale bars are 500 µm. f Sanger sequencing traces showing induction of KRASG12S and KRASG12N mutations upon using SpCas9-NG in multiplexing five mutations common in colorectal cancer tumorigenesis. The asterisk highlights the edited residue. g Quantification of editing efficiency of our 5 sgRNA CBE multiplexing experiment utilizing evolved Cas9 variant SpCas9-NG. h Quantification of single, double, triple, quadruple, and quintuple mutants acquired from a single transfection experiment in human intestinal organoids using SpCas9-NG CBE. Hom homozygous, het heterozygous, WT wild type. Source data are provided as a Source data file.