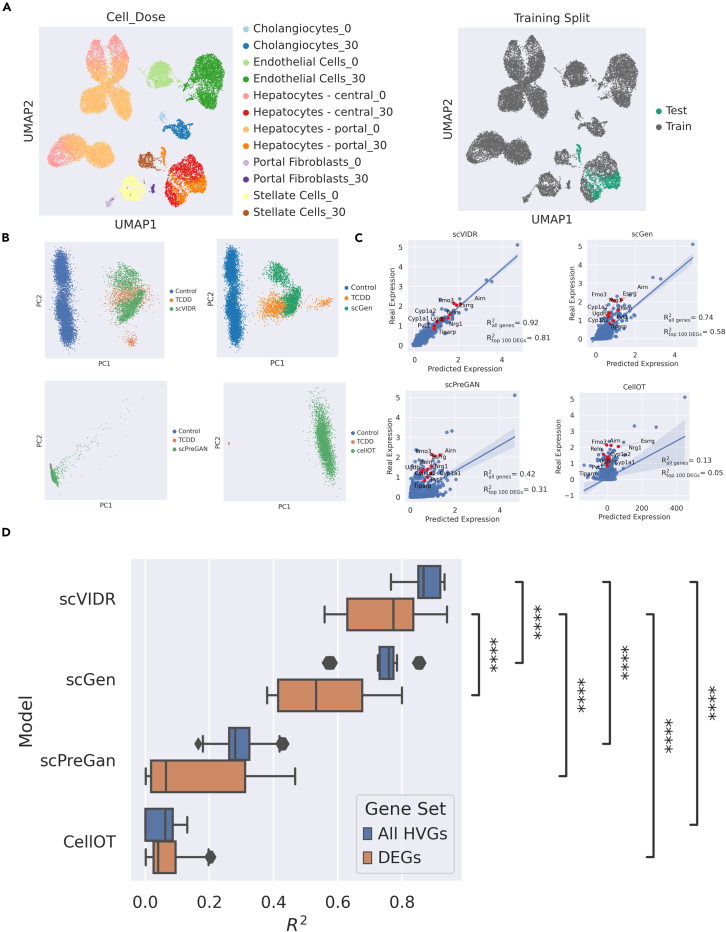
Figure 2.
Prediction of in vivo single-cell gene expression of portal hepatocytes from mice treated with 30 g/kg TCDD
(A) Uniform manifold approximation and projection (UMAP) of the latent space representation of control and treated single-cell gene expression. Each cell type and dose in g/kg combination and by the train-test split for model training is represented by different colors. In the example in the figure, TCDD-treated portal hepatocytes were used as a test set.
(B) PCA plots of predicted portal hepatocyte responses following treatment with 30 μg/kg TCDD using scGen, scVIDR, scPreGAN, and CellOT.
(C) Regression plots of each model. Each point represents the mean expression of a particular gene. Red points represent the top ten differentially expressed genes. Shaded region around regression line represents the 95% confidence interval.
(D) Boxplot of values for predictions across all liver cell types treated with 30 μg/kg TCDD. Calculation of the mean across all highly variable genes (blue). Calculation of the mean across the top 100 differentially expressed highly variable genes (orange). Prediction performance distributions were compared using a one-sided Mann-Whitney U test. ∗∗∗∗p ≤ 0.0001.