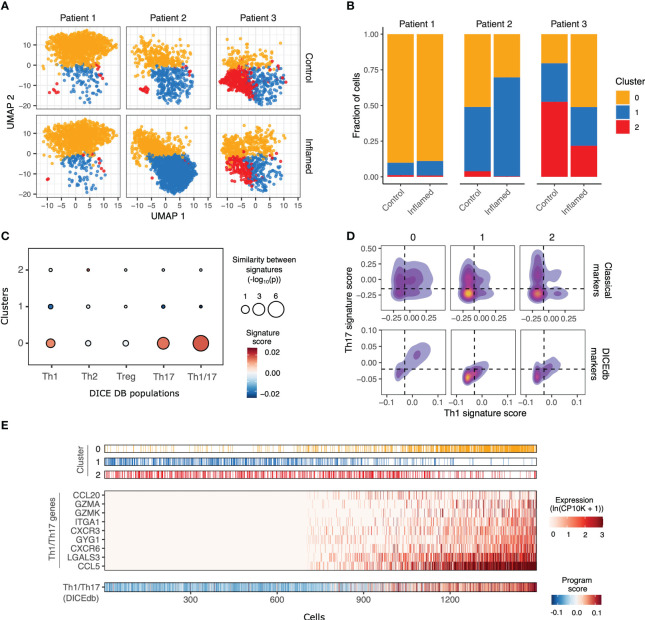
Figure 4.
Single cell transcriptomics reveals a high proportion of cytotoxic pTh17-like cells in inflamed intestinal sites of Crohn’s disease patients. (A) UMAP plots of CD4 T cells from control and inflamed samples across 3 IBD patients, colored based on Louvain clustering (C0 – C2). (B) Bar chart showing the proportion of cells from each sample mapping to each cluster. (C) Bubble plot depicts the similarity between C0-C2 cells and reference T CD4 populations from the DICEdb. Bubble size is proportional to the significance of overlap (hypergeometric test) between gene signatures from C0-C2 and DICEdb populations. Bubbles are colored according to the average expression of DICEdb signatures by cell in C0-C2. (D) Density plots showing the expression of Th1 and Th17 gene signatures by cells in C0-C2. Shown are both classical signatures (Th1: TNF, STAT1, STAT4, CXCR3 and IFNG; Th17: CCR6, STAT3, RORA, IL17A, RORC) as well as those derived from the DICEdb populations. (E) Expression of DICEdb Th1/Th17 genes by C0-C2 cells. We only included genes with significantly increased expression in C0. Cells are ordered by their Th1/Th17 scores, as shown in the lower panel. Bar on the bottom shows the expression of the complete Th1/Th17 signature by each cell. Bars on top annotate the cluster of each cell.