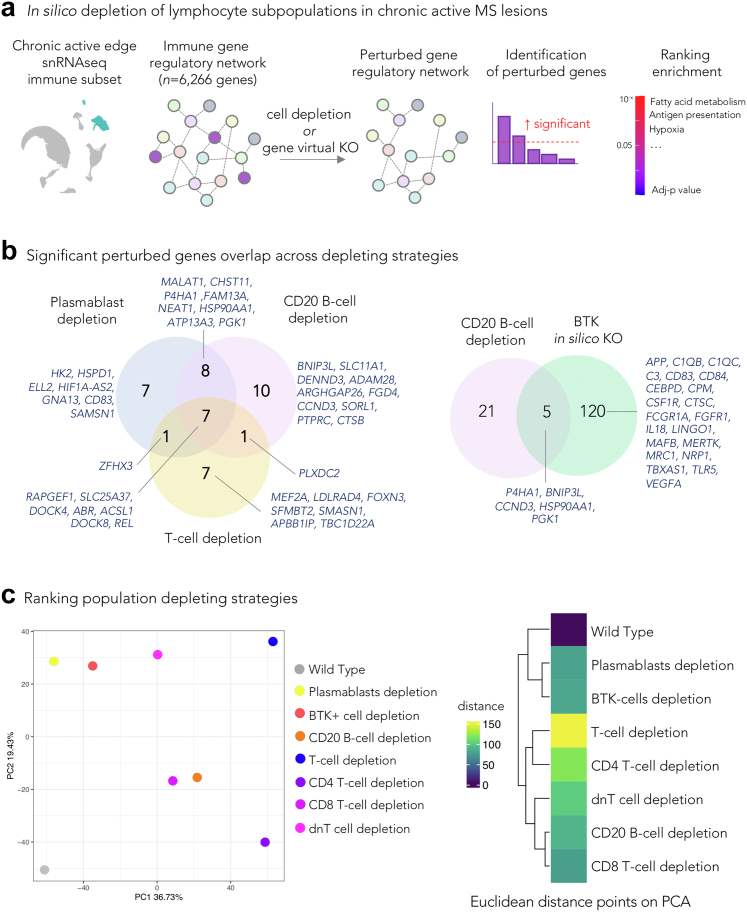
Fig. 2.
Prediction of the effects of depleting lymphocyte subpopulations in chronic active lesions. (a) Schematic workflow: starting from a snRNAseq dataset of the chronic active MS lesion edge, immune GRNs were computed and compared with GRN versions depleted of specific cell populations or of a gene of interest, using a machine learning tool for comparative single-cell network analysis (scTenifoldNet20 for comparing the effect of cell removal and scTenifoldKnk21 for the virtual knockout). The simulated significantly differentially perturbed features (genes) in the GRN comparison were analyzed for functional enrichment analysis. (b) Venn diagrams showing overlaps of significantly affected genes by in silico lymphocyte or BTK depletion. Most of these genes are known to regulate microglia or dendritic cell activities. (c) PCA plot and heatmap of the simulated distances from comparing the GRN of the original immune dataset vs the same dataset after removing specific cell populations. Wild type is the product of comparing the original dataset with itself using scTenifoldNet. This comparison serves as a reference for a null difference. The result suggests that the removal of T cells (followed by CD4 T-cells) should have a larger impact on the immune GRN at the chronic active lesion edge than removing other lymphocytes subpopulations or inhibiting BTK. The value in the tile of the heatmap is the Euclidean distance among the different comparisons; all distances are relative to the wild type comparison. The result supports the one presented in the PCA. Abbreviations: GRN: Gene regulatory network; BTK: Bruton's tyrosine kinase; dnT: CD4- CD8-double negative T cells; PCA: Principal component analysis.