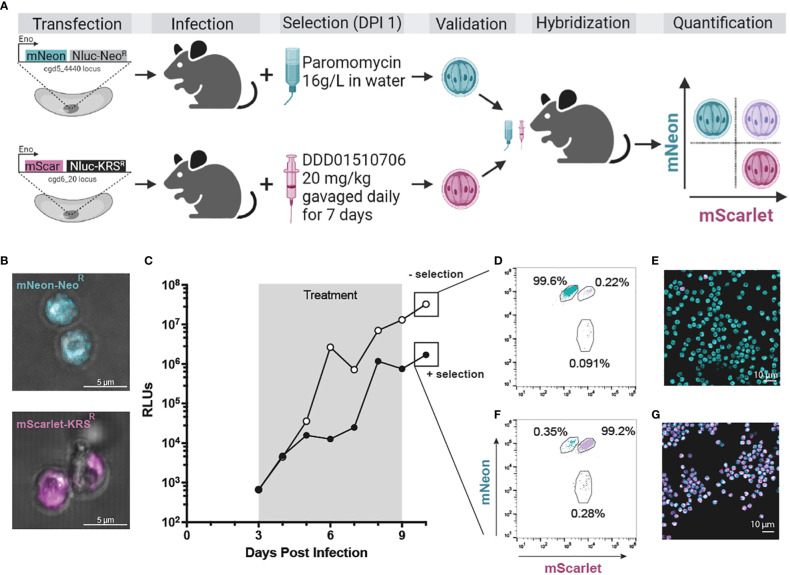
Figure 4.
KRSR is a new selection marker for Cryptosporidium genetic modification and hybridization. (A) CpTK locus was targeted for replacement with mNeon-NeoR (teal; see Supplemental Figure 3C for CRISPR strategy). In a second strain CpIMPDH was targeted for replacement with mScarlet-KRSR (magenta; see Supplemental Figure 7B for CRISPR strategy). Wild type sporozoites were transfected with either cassette and used to infect IFN-γ KO mice. Selection of transgenics began day 1 post infection (paromomycin in the water bottle or DDD01510706 by gavage, respectively; see Supplemental Figure 8 for in vivo infection data). Stable transgenic strains were obtained (Supplemental Figure 7D; Supplemental Figure 3F) and utilized in a genetic crossing experiment (microscopy of oocysts of both strains (B). Only hybrid parasites survive selection with both selection agents. Oocysts resulting from a genetic cross were quantified by flow cytometry and microscopy. An equal number of purified mNeon-NeoR and mScarlet-KRSR oocysts were used to infect two cages of mice. (C) One cage was treated with both selection agents (“+ selection”, grey box); control cage was treated with vehicle (“- selection”). Fecal samples were collected daily and infection measured by NanoLuciferase (RLUs, relative luminescence units; RLUs/2 mg of fecal material plotted). Oocysts were purified from fecal samples collected day 10 post infection. (D, E) Flow cytometry (50,000 events recorded). Flow cytometry was used to quantify the percentage of fluorescent oocysts from both selected and non-selected parasite population that express mNeon, mScarlet, or both (purple). (E, G) Purified oocysts were also analyzed by microscopy. Created with BioRender.com.