FIG. 11.
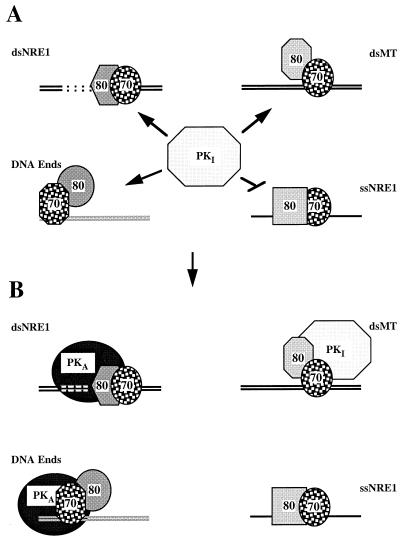
Schematic summary of the association of Ku and DNA-PKcs with the DNAs employed in this study. (A) In solution, DNA-PKcs occurs in an inactive conformation (PKI) that is attracted to Ku bound to dsNRE1 (upper left), dsMT (upper right), and double-stranded DNA ends (lower left). It does not appear to interact with Ku bound to the single, upper strand of NRE1 (ssNRE1) (lower right). The association of Ku with each of these DNAs is reflected by differences in Ku conformation, DNA conformation, and Ku-DNA contacts. In this pictogram, differences in conformation of the Ku heterodimer on DNA, as reflected by differences in protease sensitivity in EMSA and crosslinking experiments, are illustrated through differences in the shape of the Ku subunits. The Mg2+-dependent structural transition upstream of NRE1 is indicated by the dotted lines in the absence of definitive information on the exact structure of the DNA. The relative positioning of the Ku subunits with respect to the DNAs has been assigned arbitrarily. The DNA bound by Ku at the end is shown in grey to illustrate that it is not known whether a transition in DNA end structure is also important for the activation of DNA-PKcs from DNA ends. (B) The association of DNA-PKcs with Ku and DNA at NRE1 and DNA ends induces an allosteric change in the kinase that activates catalytic activity (PKA). By contrast, when associated with Ku on the MT element, DNA-PKcs remains in an inactive conformation. The positioning of DNA-PKcs over the structural transition upstream of NRE1 reflects our anticipation that this change in DNA structure may be in some way directly important for activation of the kinase at NRE1. The positioning of DNA-PKcs at the double-stranded DNA end, with Ku70 moved internally on the DNA, reflects the findings and model of Hammarsten and Chu for the assembly of Ku on DNA ends (38), with the refinement that Ku80 may not directly contact the DNA.