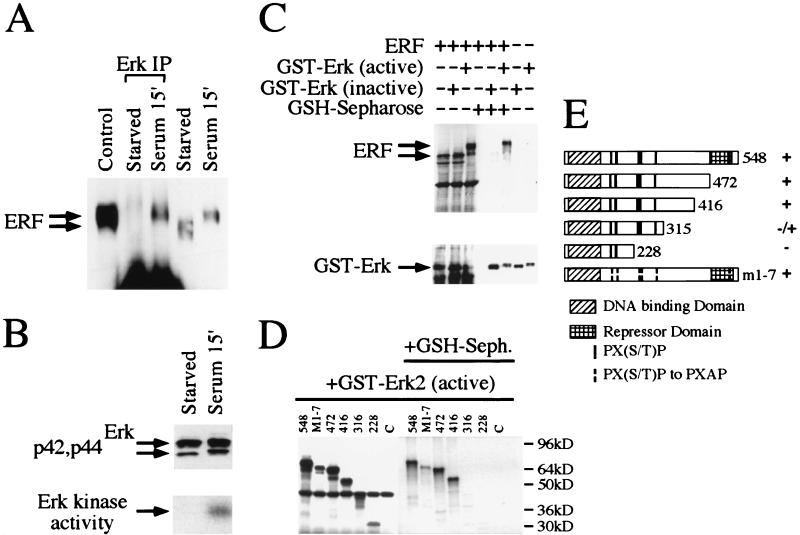
FIG. 2.
Physical interaction of ERF and Erk in vivo and in vitro. (A) Ref-1 cells were grown in the absence of serum for 20 h (starved) and then were induced for 15 min with 20% fetal calf serum (serum). Five hundred micrograms of cellular extracts from Ref-1 cells expressing ERF was immunoprecipitated with an anti-Erk rabbit polyclonal antibody under nondenaturing conditions (Erk IP) and analyzed by SDS gel electrophoresis. Twenty micrograms of cellular extracts before the addition of the anti-Erk antibody and of extracts from cells growing in complete medium (control) was also analyzed on the same gel. The presence of the ERF protein was detected by immunoblotting with the S17S anti-ERF specific antibody and is indicated by arrows. (B) Erk presence and activity in the same extracts were detected by immunoblotting (upper panel) and in-gel kinase assay (lower panel) as described for Fig. 1. (C) In vitro-translated ERF was mixed with active or inactive GST-Erk2 (Upstate Biotechnology Inc.), and the complexes were precipitated with GSH-Sepharose and analyzed by SDS gel electrophoresis. The presence or absence of ERF was detected by autoradiography (upper panel), and that of GST-Erk2 was detected by immunoblotting with an anti-Erk specific antibody (lower panel). (D) The presence or absence of truncated ERF proteins in complex with active GST-Erk2 was detected as described for panel C. The numbers indicate the carboxy-terminal amino acid of each deletion. M1-7 is a full-length ERF with all seven putative MAPK sites mutated to alanine. (E) Diagrammatic representation of the deletions and mutation used for panel D. The Ets DNA-binding domain, the previously identified repression domain, and the putative MAPK phosphorylation sites are indicated. Plus and minus indicate the interaction of each protein with active GST-Erk2 kinase.