Abstract
Phantom tooth pain (PTP) is a rare and specific neuropathic pain that occurs after pulpectomy and tooth extraction, but its cause is not understood. We hypothesized that there is a genetic contribution to PTP. The present study focused on the CACNA1C gene, which encodes the α1C subunit of the Cav1.2 L-type Ca2+ channel (LTCC) that has been reported to be associated with neuropathic pain in previous studies. We investigated genetic polymorphisms that contribute to PTP. We statistically examined the association between genetic polymorphisms and PTP vulnerability in 33 patients with PTP and 118 patients without PTP but with pain or dysesthesia in the orofacial region. From within and around the CACNA1C gene, 155 polymorphisms were selected and analyzed for associations with clinical data. We found that the rs216009 single-nucleotide polymorphism (SNP) of the CACNA1C gene in the recessive model was significantly associated with the vulnerability to PTP. Homozygote carriers of the minor C allele of rs216009 had a higher rate of PTP. Nociceptive transmission in neuropathic pain has been reported to involve Ca2+ influx from LTCCs, and the rs216009 polymorphism may be involved in CACNA1C expression, which regulates intracellular Ca2+ levels, leading to the vulnerability to PTP. Furthermore, psychological factors may lead to the development of PTP by modulating the descending pain inhibitory system. Altogether, homozygous C-allele carriers of the rs216009 SNP were more likely to be vulnerable to PTP, possibly through the regulation of intracellular Ca2+ levels and affective pain systems, such as those that mediate fear memory recall.
Keywords: phantom tooth pain, voltage-dependent calcium channel, L-type calcium channel, Cav1.2, single-nucleotide polymorphism, neuropathic pain
Introduction
Advanced dental caries causes the infection of dental pulp, for which the removal of dental pulp (i.e., pulpectomy) is required. There is usually no residual pain after pulpectomy, but pain can occasionally occur. Pain may also occur in the same area after tooth extraction. The rare pain that occurs after pulpectomy or tooth extraction is known as phantom tooth pain (PTP), a type of specific neuropathic pain. 1 Although Melzack’s neuromatrix theory may be relevant because of its essentially similar characteristics to phantom limb pain after limb amputation, 1 the cause of PTP remains unclear. In phantom limb pain, genetic factors have been reported in animal studies. 1 In PTP in humans, Soeda et al. showed that the rs735055 single-nucleotide polymorphism (SNP) of the SLC17A9 gene and rs3732759 SNP of the P2RY12 gene are associated with the development of PTP, 2 but little is known about other genetic factors.
Nociceptive stimuli and neuropathy generate pain through various receptors and ion channels. One such channel is the calcium (Ca2+) channel. Pain stimulation increases intracellular Ca2+, resulting in intracellular signaling. The generation and transmission of pain involve the action of voltage-dependent Ca2+ channels (VDCCs). VDCCs are classified into two main types: high voltage-activated and low voltage-activated. High voltage-activated VDCCs consist of a heterotetramer that is composed of α1, α2δ, β, and γ subunits. The α1 subunit protein is encoded by 10 different genes that are classified into L-type (Cav1), P/Q-type (Cav2.1), N-type (Cav2.2), R-type (Cav2.3), and T-type (Cav3) according to their specific characteristics. L-type Ca2+ channels (LTCCs) are known to regulate the activity of transcription factors by working in concert with enzymes that are involved in phosphorylation (which is important for gene expression), the contraction of skeletal, cardiac, and smooth muscles, and the release of hormones and neurotransmitters.3–5
Central sensitization may be maintained for several days after the pain stimulus has ceased, which is associated with symptoms of allodynia. 6 Two LTCCs, Cav1.2 and Cav1.3, are present in the dorsal horn of the spinal cord. Cav1.2 channels play a minor role in central sensitization, but they are known to be associated with neuropathic pain by modulating gene expression regulating intracellular Ca2+ influx.
Most patients with PTP meet the criteria for somatoform pain disorders in the Diagnostic and Statistical Manual of Mental Disorders, 5th edition (DSM-5), and are often referred to as having nociplastic pain. 1 Human genetic variants of the CACNA1C gene, which encodes the Cav1.2 α1C subunit protein (CACNA1C), are widely associated with a higher risk of neuropsychiatric disorders, including depression, bipolar disorder, and schizophrenia.7,8 Cav1.2 LTCCs have also been reported to be involved in affective pain systems, such as social fear learning and fear memory recall, in the anterior cingulate cortex (ACC). 9 Based on these reports, we hypothesized that the CACNA1C gene in the ACC region is involved in the development of PTP through psychogenic factors.
We postulated that the genetic cause of neuropathic PTP may involve SNPs of the CACNA1C gene. We statistically analyzed differences in gene polymorphism frequencies between patients with PTP (i.e., neuropathic pain in the oral and maxillofacial regions) and other patients without PTP but with pain or dysesthesia in the orofacial region (i.e., orofacial pain [OFP]). The results showed a significant association between the rs216009 SNP of CACNA1C and the vulnerability to PTP.
Materials and Methods
Patients
The present study was approved by the Ethics Committees of Tokyo Dental College and Tokyo Metropolitan Institute of Medical Science (approval no. 810 and 20-45, respectively). The study was performed in accordance with provisions of the Declaration of Helsinki. All subjects provided written informed consent for the genetics studies.
The study enrolled 33 PTP patients (26-74 years old) and 118 patients without PTP but with pain or dysesthesia in the orofacial region (OFP; 23-89 years old) who visited Tokyo Dental College Suidobashi Hospital from May 2007 to November 2019. The patients were classified as traumatic trigeminal neuropathy, trigeminal neuralgia, postherpetic neuralgia, neuralgia-inducing cavitational osteonecrosis, and nociplastic pain based on the International Classification of Orofacial Pain, 1st edition (ICOP), 10 and International Statistical Classification of Diseases and Related Health Problems, 11th revision (ICD-11). 11 Sixty patients had traumatic trigeminal neuropathy (39 patients experienced no pain, 21 patients experienced pain), 11 patients had trigeminal neuralgia, 17 patients had postherpetic neuralgia, 12 patients had neuralgia-inducing cavitational osteonecrosis (NICO), and 18 patients had nociplastic pain. The difference between nociplastic pain and neuropathic pain remains unclear. Therefore, the classification is unclear even in the ICD-11 and ICOP.10,11 The following definitions were added in this study to clarify these differences. The following items were applied as diagnostic criteria for PTP: (1) allodynia in surrounding gingiva after pulp extraction and presence of pain that is unresponsive to local infiltration anesthesia and (2) post-extraction pain with residual pain despite good healing of the mucous that covers the tooth, the presence of allodynia, and pain that is unresponsive to local infiltration anesthesia. 2
Genotyping and linkage disequilibrium analysis
We examined SNPs of the CACNA1C gene. The genotype data from the whole-genome genotyping of 151 patients with PTP or OFP were used to analyze 303 SNPs within and around the CACNA1C gene region (including 10 kilobase pairs [kbp] upstream and downstream). Genomic DNA was extracted from whole blood samples using standard procedures. The extracted DNA was dissolved in TE buffer (10 mM Tris-HCl and 1 mM ethylenediaminetetraacetic acid, pH 8.0). Whole-genome genotyping was performed after measuring DNA concentrations using a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Tokyo, Japan), with the concentration adjusted to 100 ng/μl. Whole-genome genotyping was performed using the Infinium Assay II and iScan system (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Infinium Asian Screening Array-24 v1.0 BeadChips (total markers: 659,184) were used for genotyping in the genetic analysis. The BeadChips also contained several probes that were specific to copy number variation markers, but most target SNP markers on human autosomal or sex chromosomes. Data from whole-genome-genotyped samples were extracted and analyzed using GenomeStudio 2.0 with Genotyping module v3.3.7 to assess quality of the results for SNPs within and around the CACNA1C gene region. To include flanking regions of the gene, SNPs were selected within a range of 10 kbp each upstream and downstream of the CACNA1C gene region. During the data cleaning process, samples with genotyping rates less than 0.95 were excluded from further analysis. As a result, no samples were excluded from further analysis. Markers with genotype call frequencies less than 0.95 or “cluster segregation” (i.e., a measure of genotype cluster segregation) less than 0.1 were excluded from the subsequent association studies. A total of 303 SNP markers survived the filtering process for this patient sample for the region that was investigated. Linkage disequilibrium (LD) analysis was performed on 303 SNPs in the CACNA1C gene region in the SNP array. The 148 SNPs with minor allele frequencies less than 0.05 were excluded from the LD analysis, and the remaining 155 SNPs were employed for further analysis. To estimate LD intensity between SNPs, the commonly used D' and r 2 values were calculated pairwise using the genotype dataset for each SNP. The LD block was defined among SNPs that showed “strong LD” based on the default algorithm of Gabriel et al. with an upper limit of 0.98 and a lower limit of 0.7 for the 95% confidence interval of D' that indicated strong LD. TagSNPs in the LD blocks were determined using the Tagger software package that is incorporated in Haploview, which was detailed in a previous report. 12
Statistical analysis
For all genotype frequency data, deviations from the theoretical Hardy-Weinberg equilibrium distribution were examined, and χ 2 tests were performed to analyze associations with clinical data for PTP. The χ 2 tests were performed using SPSS 28 software (IBM Japan, Tokyo, Japan). For all statistical tests, the criterion for significance was p < .05. Bonferroni correction for multiple comparisons was performed for 155 SNPs in the genotypic, dominant, and recessive models for each minor allele.
Results
CACNA1C gene rs216009 SNP was associated with PTP
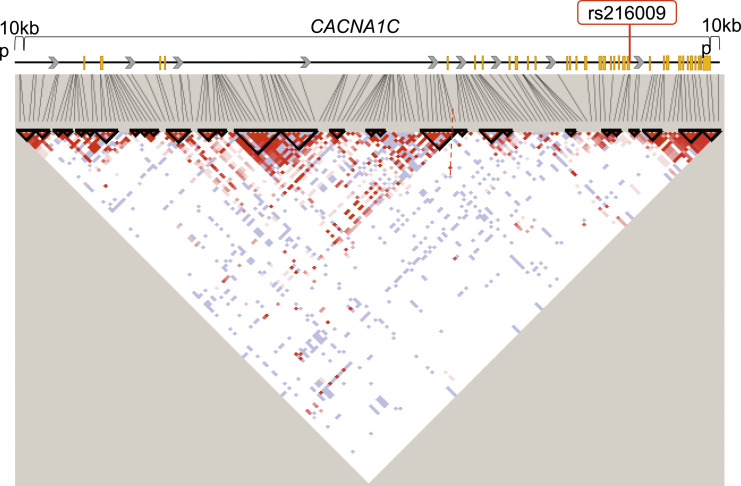
We focused on the CACNA1C gene, which is involved in neuropathic pain. The SNPs within and around the CACNA1C gene that were extracted from whole-genome genotyping data from PTP and OFP patients and the subsequent LD analysis resulted in the selection of 73 TagSNPs and a total of 29 LD blocks. D' and r 2 values are presented in Supplemental Table S1. A schematic diagram of the CACNA1C gene and r 2 values is presented in Figure 1. The genotype data consisted of three genotypes. In the genotypic and dominant models, the results were not significant for any SNPs (corrected p > .05) after Bonferroni correction for multiple comparisons (Supplemental Table S2, S3). In the recessive model, the results were significant only for the rs216009 SNP (corrected p = 4.1 × 10−2) after Bonferroni correction for multiple comparisons (Table 1). The rs216009 SNP did not deviate from theoretical Hardy-Weinberg equilibrium (Supplemental Table S4). Based on these results, the association between the rs216009 SNP of the CACNA1C gene and PTP was significant in the recessive model. An enlarged view of the area around the rs216009 polymorphism in Figure 1 is shown in Supplemental Figure S1. There was a higher rate of C-allele homozygote carriers in the PTP group than in the OFP group (PTP: CC/total = 39% [13/33], OFP: CC/total = 12% [14/118]). C-allele homozygote carriers had a higher incidence of PTP, suggesting that the C allele of the rs216009 SNP of the CACNA1C gene is associated with a higher risk of PTP in an autosomal recessive manner.
Figure 1.
State of linkage disequilibrium (LD) among SNPs in the CACNA1C gene region, including 10 kbp upstream and downstream (LD Plot-r2). D' and r 2 values are presented in Supplemental Table S1, S3. White boxes represent D' < 1, log of likelihood odds ratio (LOD) < 2. Pink and red boxes represent D' < 1 and LOD ≥ 2. Blue boxes represent D' = 1 and LOD < 2. Bright red boxes represent D' = 1 and LOD ≥ 2. The solid horizontal line above the LD plot represents the CACNA1C gene, including 10 kbp upstream and downstream of the gene. The yellow boxes in the structure of the CACNA1C gene represent exons, and the solid lines represent untranslated regions or introns. The gray arrows represent the direction of transcription. The red squares represent the SNP of interest in this study.
Table 1.
Results of association analysis of CACNA1C SNPs in the recessive model.
| SNP | Alleles | Genotypes (Recessive) | AFF a | UNAFF b | χ 2 | P-value | P c -value | BP |
|---|---|---|---|---|---|---|---|---|
| Minor | Major | |||||||
| kgp12074188 | G | A | 1/32 | 6/112 | 0.2462 | 0.6198 | 1 | 1966486 |
| 12:2075984 | C | T | 1/32 | 6/112 | 0.2462 | 0.6198 | 1 | 1966818 |
| rs11062040 | C | T | 7/26 | 23/95 | 0.04795 | 0.8267 | 1 | 1982091 |
| kgp9386543 | A | G | 0/33 | 0/118 | NA | NA | NA | 1983811 |
| rs2041135 | C | T | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 1992332 |
| kgp3992595 | C | T | 7/26 | 23/95 | 0.04795 | 0.8267 | 1 | 2006081 |
| rs3858698 | T | C | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2006595 |
| rs2429127 | C | T | 1/32 | 5/113 | 0.09846 | 0.7537 | 1 | 2007602 |
| kgp12186303 | A | G | 0/33 | 0/118 | NA | NA | NA | 2021273 |
| rs61481868 | T | C | 7/26 | 19/99 | 0.4725 | 0.4918 | 1 | 2023329 |
| rs4765876 | C | A | 1/32 | 0/118 | 3.6 | 0.05779 | 1 | 2023935 |
| kgp9460814 | A | G | 1/32 | 0/118 | 3.6 | 0.05779 | 1 | 2025860 |
| kgp11189985 | T | C | 3/30 | 3/115 | 2.898 | 0.08867 | 1 | 2026321 |
| kgp1012928 | A | G | 7/26 | 12/106 | 2.859 | 0.09087 | 1 | 2026751 |
| kgp5132388 | G | A | 5/28 | 3/115 | 8.172 | 0.004255 | 0.659525 | 2028978 |
| kgp10845951 | T | C | 0/33 | 2/116 | 0.5668 | 0.4515 | 1 | 2031691 |
| GSA-rs917365 | G | A | 1/32 | 4/114 | 0.01041 | 0.9187 | 1 | 2043005 |
| rs9888329 | G | A | 2/31 | 1/117 | 3.599 | 0.05781 | 1 | 2044068 |
| rs723672 | C | T | 1/32 | 2/116 | 0.2362 | 0.627 | 1 | 2052395 |
| JHU_12.2161641 | C | T | 8/25 | 16/102 | 2.202 | 0.1379 | 1 | 2052476 |
| kgp7268544 | G | A | 2/31 | 1/117 | 3.599 | 0.05781 | 1 | 2057313 |
| rs11062093 | G | A | 2/31 | 1/117 | 3.599 | 0.05781 | 1 | 2057313 |
| rs2215095 | A | G | 2/31 | 1/117 | 3.599 | 0.05781 | 1 | 2057872 |
| kgp5897423 | A | G | 0/33 | 0/118 | NA | NA | NA | 2061727 |
| rs57860259 | G | A | 1/32 | 0/118 | 3.6 | 0.05779 | 1 | 2062463 |
| rs2370251 | C | T | 8/25 | 18/100 | 1.462 | 0.2267 | 1 | 2063125 |
| kgp12182728 | G | A | 4/29 | 4/114 | 3.918 | 0.04776 | 1 | 2066137 |
| kgp11173803 | A | G | 3/30 | 5/113 | 1.211 | 0.2712 | 1 | 2068380 |
| kgp567525 | A | G | 3/30 | 5/113 | 1.211 | 0.2712 | 1 | 2069348 |
| rs2283271 | A | T | 5/28 | 16/102 | 0.0546 | 0.8152 | 1 | 2073132 |
| kgp4327291 | T | C | 1/32 | 6/111 | 0.2546 | 0.6138 | 1 | 2079543 |
| rs2283277 | A | G | 2/31 | 5/113 | 0.1939 | 0.6597 | 1 | 2084670 |
| rs2283280 | C | A | 4/29 | 13/105 | 0.03147 | 0.8592 | 1 | 2099224 |
| rs758723 | A | T | 9/24 | 17/100 | 2.917 | 0.08765 | 1 | 2111239 |
| rs2238032 | G | T | 1/32 | 1/117 | 0.9402 | 0.3322 | 1 | 2113566 |
| rs2238034 | C | T | 3/30 | 9/109 | 0.07553 | 0.7834 | 1 | 2126462 |
| 12:2245488 | A | G | 0/33 | 2/116 | 0.5668 | 0.4515 | 1 | 2136322 |
| rs12579529 | G | A | 1/32 | 1/117 | 0.9402 | 0.3322 | 1 | 2136470 |
| 12:2253686 | A | G | 0/33 | 0/118 | NA | NA | NA | 2144520 |
| kgp3137218 | T | G | 6/27 | 16/102 | 0.4427 | 0.5058 | 1 | 2161776 |
| kgp5525565 | A | G | 7/26 | 27/91 | 0.04119 | 0.8392 | 1 | 2164749 |
| rs10848622 | G | A | 5/28 | 21/97 | 0.1266 | 0.722 | 1 | 2164885 |
| rs74062239 | G | A | 0/33 | 2/116 | 0.5668 | 0.4515 | 1 | 2165753 |
| rs11062138 | A | G | 11/22 | 23/95 | 2.832 | 0.0924 | 1 | 2171776 |
| imm_12_2153456 | C | T | 1/32 | 6/112 | 0.2462 | 0.6198 | 1 | 2174029 |
| kgp1967023 | A | G | 5/28 | 14/104 | 0.2533 | 0.6147 | 1 | 2174226 |
| GSA-rs7298845 | G | A | 3/30 | 7/111 | 0.4161 | 0.5189 | 1 | 2175167 |
| imm_12_2155992 | A | G | 6/27 | 8/110 | 3.986 | 0.04589 | 1 | 2176565 |
| kgp6010347 | G | A | 2/31 | 7/111 | 0.0007586 | 0.978 | 1 | 2176818 |
| rs74238853 | A | G | 1/32 | 1/117 | 0.9402 | 0.3322 | 1 | 2177931 |
| kgp1860328 | C | T | 6/27 | 8/110 | 3.986 | 0.04589 | 1 | 2179239 |
| imm_12_2158697 | C | T | 6/27 | 8/110 | 3.986 | 0.04589 | 1 | 2179270 |
| imm_12_2162951 | A | G | 7/26 | 9/109 | 5.024 | 0.025 | 1 | 2183524 |
| rs2238051 | C | A | 4/28 | 15/103 | 0.001021 | 0.9745 | 1 | 2198009 |
| imm_12_2186878 | C | T | 4/29 | 6/112 | 2.065 | 0.1507 | 1 | 2207451 |
| imm_12_2201258 | A | C | 4/29 | 6/112 | 2.065 | 0.1507 | 1 | 2221831 |
| GSA-rs11062162 | G | A | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2222938 |
| kgp7118789 | A | G | 2/31 | 5/113 | 0.1939 | 0.6597 | 1 | 2227972 |
| kgp5423214 | A | G | 4/29 | 5/113 | 2.86 | 0.09082 | 1 | 2233082 |
| imm_12_2212678 | G | A | 4/29 | 5/113 | 2.86 | 0.09082 | 1 | 2233251 |
| rs2007044 | G | A | 5/28 | 7/111 | 2.996 | 0.08346 | 1 | 2235794 |
| rs1006737 | A | G | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2236129 |
| rs4765905 | C | G | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2240418 |
| rs7975467 | G | A | 3/30 | 6/112 | 0.7384 | 0.3902 | 1 | 2241097 |
| rs7297582 | T | C | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2246640 |
| rs2283292 | A | G | 3/30 | 4/114 | 1.896 | 0.1685 | 1 | 2262486 |
| rs4765913 | A | T | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2310730 |
| kgp49187 | G | A | 4/29 | 16/102 | 0.04641 | 0.8294 | 1 | 2311078 |
| kgp4177886 | A | G | 7/26 | 17/101 | 0.8934 | 0.3446 | 1 | 2312422 |
| rs2283295 | A | G | 1/32 | 0/118 | 3.6 | 0.05779 | 1 | 2314745 |
| rs3819536 | A | G | 8/25 | 26/92 | 0.0721 | 0.7883 | 1 | 2327832 |
| rs2239050 | C | G | 1/32 | 0/118 | 3.6 | 0.05779 | 1 | 2338248 |
| 12:2450156 | A | G | 1/32 | 1/117 | 0.9402 | 0.3322 | 1 | 2340990 |
| kgp3584318 | C | T | 8/25 | 25/93 | 0.141 | 0.7073 | 1 | 2354572 |
| kgp11928887 | G | A | 0/33 | 2/116 | 0.5668 | 0.4515 | 1 | 2354579 |
| rs61294626 | G | A | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2354848 |
| kgp886037 | C | T | 0/33 | 0/118 | NA | NA | NA | 2355523 |
| kgp11813198 | T | C | 6/27 | 16/102 | 0.4427 | 0.5058 | 1 | 2356311 |
| rs2238077 | A | G | 1/32 | 9/109 | 0.8812 | 0.3479 | 1 | 2356918 |
| rs80004763 | T | C | 1/32 | 0/118 | 3.6 | 0.05779 | 1 | 2359192 |
| kgp4449116 | A | C | 1/32 | 0/118 | 3.6 | 0.05779 | 1 | 2360201 |
| kgp944949 | A | G | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2360392 |
| kgp1280039 | G | A | 2/31 | 3/114 | 0.9766 | 0.323 | 1 | 2362100 |
| rs11062202 | A | G | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2363095 |
| JHU_12.2476637 | T | C | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2367472 |
| kgp7855943 | T | C | 1/32 | 7/111 | 0.4328 | 0.5106 | 1 | 2368307 |
| kgp12453198 | A | G | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2369824 |
| rs886898 | A | G | 2/31 | 8/110 | 0.02156 | 0.8833 | 1 | 2372770 |
| kgp12240640 | G | A | 7/26 | 24/94 | 0.01205 | 0.9126 | 1 | 2385760 |
| kgp6704767 | A | G | 1/32 | 6/112 | 0.2462 | 0.6198 | 1 | 2387667 |
| JHU_12.2506037 | A | G | 2/31 | 2/116 | 1.906 | 0.1674 | 1 | 2396872 |
| rs2239063 | C | A | 3/30 | 12/106 | 0.03353 | 0.8547 | 1 | 2402665 |
| rs11831085 | G | A | 2/31 | 2/116 | 1.906 | 0.1674 | 1 | 2405692 |
| rs10774042 | C | T | 2/31 | 15/103 | 1.142 | 0.2853 | 1 | 2409963 |
| rs116846988 | T | C | 0/33 | 2/116 | 0.5668 | 0.4515 | 1 | 2411192 |
| kgp783053 | G | A | 1/32 | 7/111 | 0.4328 | 0.5106 | 1 | 2414090 |
| rs7312105 | G | A | 2/31 | 14/104 | 0.9169 | 0.3383 | 1 | 2414189 |
| rs2239073 | T | C | 3/30 | 13/105 | 0.101 | 0.7507 | 1 | 2429334 |
| rs2239074 | T | C | 2/31 | 4/114 | 0.4821 | 0.4875 | 1 | 2429383 |
| JHU_12.2541533 | C | T | 0/33 | 0/118 | NA | NA | NA | 2432368 |
| kgp3996071 | G | A | 4/29 | 21/97 | 0.6012 | 0.4381 | 1 | 2433934 |
| kgp3964892 | G | A | 6/27 | 12/106 | 1.577 | 0.2092 | 1 | 2436413 |
| rs17801211 | A | G | 5/28 | 17/101 | 0.01149 | 0.9146 | 1 | 2437576 |
| kgp8502710 | A | G | 0/33 | 3/115 | 0.856 | 0.3549 | 1 | 2439108 |
| rs1860102 | A | G | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2439324 |
| rs1076344 | C | T | 1/32 | 0/118 | 3.6 | 0.05779 | 1 | 2444822 |
| rs2239080 | G | A | 3/30 | 20/98 | 1.233 | 0.2668 | 1 | 2445523 |
| rs7303824 | T | C | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2448839 |
| kgp11101662 | A | C | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2451998 |
| 12:2561715 | G | A | 2/31 | 17/101 | 1.633 | 0.2013 | 1 | 2452549 |
| rs10774048 | T | C | 0/33 | 3/115 | 0.856 | 0.3549 | 1 | 2453375 |
| kgp5800475 | C | T | 1/32 | 4/114 | 0.01041 | 0.9187 | 1 | 2454249 |
| kgp6818367 | T | C | 1/32 | 6/112 | 0.2462 | 0.6198 | 1 | 2464879 |
| kgp387637 | A | G | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2474402 |
| kgp2586442 | C | T | 3/30 | 6/112 | 0.7384 | 0.3902 | 1 | 2481863 |
| JHU_12.2598802 | C | T | 1/32 | 1/117 | 0.9402 | 0.3322 | 1 | 2489637 |
| rs16929471 | A | G | 1/32 | 0/118 | 3.6 | 0.05779 | 1 | 2492577 |
| kgp1764861 | A | G | 0/33 | 0/118 | NA | NA | NA | 2499317 |
| JHU_12.2608616 | A | G | 4/29 | 16/102 | 0.04641 | 0.8294 | 1 | 2499451 |
| kgp12551081 | C | T | 0/33 | 0/118 | NA | NA | NA | 2499918 |
| kgp12044379 | A | G | 1/32 | 3/115 | 0.02381 | 0.8774 | 1 | 2500108 |
| JHU_12.2616027 | A | C | 1/32 | 1/117 | 0.9402 | 0.3322 | 1 | 2506862 |
| JHU_12.2621911 | A | G | 1/32 | 1/117 | 0.9402 | 0.3322 | 1 | 2512746 |
| kgp2427637 | T | C | 10/23 | 27/91 | 0.7678 | 0.3809 | 1 | 2513227 |
| rs2239101 | C | T | 1/31 | 1/117 | 0.9926 | 0.3191 | 1 | 2513313 |
| rs2283324 | T | C | 2/31 | 1/117 | 3.599 | 0.05781 | 1 | 2537319 |
| JHU_12.2677376 | C | A | 2/31 | 1/117 | 3.599 | 0.05781 | 1 | 2568211 |
| kgp312155 | T | C | 0/33 | 0/118 | NA | NA | NA | 2575247 |
| rs11832738 | G | A | 2/31 | 6/112 | 0.04895 | 0.8249 | 1 | 2581536 |
| rs215976 | T | C | 4/29 | 11/107 | 0.2258 | 0.6346 | 1 | 2585472 |
| rs215981 | C | T | 4/29 | 10/108 | 0.4077 | 0.5232 | 1 | 2596190 |
| rs215994 | C | T | 6/27 | 18/100 | 0.1653 | 0.6843 | 1 | 2607994 |
| rs216008 | T | C | 6/27 | 19/99 | 0.08077 | 0.7763 | 1 | 2611971 |
| kgp8181528 | A | G | 0/33 | 2/116 | 0.5668 | 0.4515 | 1 | 2613098 |
| rs216009 | C | T | 13/20 | 14/104 | 13.31 | 0.000264 | 0.04092* | 2613367 |
| rs216013 | G | A | 6/27 | 19/99 | 0.08077 | 0.7763 | 1 | 2620466 |
| kgp8035649 | A | C | 4/29 | 10/108 | 0.4077 | 0.5232 | 1 | 2623553 |
| rs35240807 | A | G | 0/33 | 1/117 | 0.2815 | 0.5957 | 1 | 2637599 |
| rs216026 | T | G | 1/32 | 7/111 | 0.4328 | 0.5106 | 1 | 2639961 |
| rs11613210 | A | C | 1/32 | 8/110 | 0.6468 | 0.4213 | 1 | 2642146 |
| rs216029 | C | T | 1/32 | 8/110 | 0.6468 | 0.4213 | 1 | 2645616 |
| rs2239128 | T | C | 2/31 | 13/105 | 0.7081 | 0.4001 | 1 | 2648603 |
| rs740728 | C | T | 1/32 | 6/112 | 0.2462 | 0.6198 | 1 | 2650137 |
| rs12312322 | A | G | 2/31 | 9/109 | 0.0937 | 0.7595 | 1 | 2662640 |
| rs2302727 | A | C | 1/32 | 2/116 | 0.2362 | 0.627 | 1 | 2665435 |
| rs7295250 | T | C | 4/29 | 16/102 | 0.04641 | 0.8294 | 1 | 2667777 |
| rs11062301 | A | G | 2/31 | 9/109 | 0.0937 | 0.7595 | 1 | 2667787 |
| rs10744567 | A | G | 11/22 | 28/90 | 1.242 | 0.2651 | 1 | 2671650 |
| rs2302729 | T | C | 11/22 | 28/90 | 1.242 | 0.2651 | 1 | 2674806 |
| rs1051375 | G | A | 11/22 | 28/88 | 1.124 | 0.289 | 1 | 2679713 |
| rs10774053 | A | G | 11/22 | 28/90 | 1.242 | 0.2651 | 1 | 2681966 |
| kgp6442815 | A | G | 2/31 | 9/109 | 0.0937 | 0.7595 | 1 | 2686468 |
| rs7316246 | A | G | 0/33 | 4/114 | 1.149 | 0.2837 | 1 | 2695289 |
| kgp4023122 | A | G | 0/33 | 4/114 | 1.149 | 0.2837 | 1 | 2698503 |
| 12:2816060 | G | A | 0/33 | 4/114 | 1.149 | 0.2837 | 1 | 2706894 |
P-values before Bonferroni correction.
NA: not available.
aNumber of samples with PTP.
bNumber of samples without PTP.
cP values after Bonferroni correction for multiple comparisons.
The corrected P value, which is P > 1, is indicated as 1.
* P < 0.05.
Discussion
The present findings suggest that the rs216009 SNP of the CACNA1C gene is significantly associated with the vulnerability to PTP. Homozygote carriers of the minor C allele of the rs216009 SNP of the CACNA1C gene were significantly more likely to be affected by PTP.
Significant differences were found between PTP and OFP. Orofacial pain comprises painless trigeminal neuropathy, nociplastic pain, and neuropathic pain other than PTP (including painful trigeminal neuropathy, trigeminal neuralgia, postherpetic neuralgia, and NICO). To clarify differences between these OFP subgroups and PTP, we statistically analyzed differences between each subgroup and PTP, although the number of people in each subgroup was small. The results (Supplemental Table S5) showed that PTP was significantly different from other neuropathic pain, nociplastic pain, and painless trigeminal neuropathy. These results suggest that PTP is specific and does not fit any diagnostic criteria that were defined by Marbach with regard to the rs216009 SNP of CACNA1C, although Marbach classified it as the same neuropathic pain. 1 This may be partially attributable to impairments in brain transmission in ACC regions and other areas. In the Genotype-Tissue Expression (GTEx) database, the rs216009 SNP is located in the peak region of H3K27ac enrichment in Brodmann area 9 (BA9) of the human prefrontal cortex (PFC), heart, and muscle (Supplemental Figure S2). 13 H3K27ac is known to be involved in enhancer activity, suggesting that the rs216009 SNP is located in the enhancer region of the CACNA1C gene in BA9 of the PFC, heart, and muscle. Additionally, the ACC is closely associated with the cerebral cortex, including the PFC (BA9), which is interconnected with areas that are important for pain processing.14–16 Cav1.2 LTCCs in the ACC are involved in observational fear learning (affective pain system). 9 The ACC is an important brain region for the convergence of sensory and emotional information and has been reported to potentially mediate emotional responses to nociceptive stimuli. The ACC has also been reported to exhibit anatomical and neurochemical changes in chronic pain patients. 16 In chronic pain and nociplastic pain, pain is enhanced by cerebral cortex activity via the periaqueductal gray (PAG)–rostroventral medulla system (the descending pain inhibitory system). 16 Therefore, one possibility is that tissue through the ACC–PFC–PAG pathway in the brain may also have enhancer activity at the rs216009 SNP site. The CACNA1C gene has been associated with various psychiatric disorders.7,8 Many patients with PTP have also been reported to meet DSM-5 diagnostic criteria for a somatoform pain disorder, often referred to as nociplastic pain. 1 The rs216009 SNP of the CACNA1C gene was shown to be significantly associated with PTP in the present study. Thus, the affective pain system may be involved in the development of CACNA1C-mediated PTP. Psychological factors may lead to the development of PTP by modulating the descending pain inhibitory system in the ACC–PFC–PAG pathway in the brain in patients who carry the homozygous C-allele of the rs216009 SNP of the CACNA1C gene. However, further research is needed to elucidate pathways that are involved in the development of PTP.
Genetic mutations of CACNA1C are also known to be a risk factor for posttraumatic stress disorder (PTSD). 17 Dopamine D1 receptors have been reported to be involved in the prolongation of remote fear memories and vulnerability to PTSD. Dopamine is involved in contextual fear memory, and Cav1.2 LTCCs are a downstream target of D1 receptor signaling. Bavley et al. used Cacna1c knockout mice to examine remote contextual fear after the onset of PTSD-like symptoms. 17 Their results suggested that Cacna1c expression inhibits fear memory recall and that Cav1.2 LTCCs may be responsible for neurogenesis in the hippocampus. Fear memories are involved in the emotional pain system, suggesting that the Cacna1c gene may be associated with the affective pain system through neurogenesis. The present study found that the rs216009 SNP of the CACNA1C gene is associated with PTP, suggesting that CACNA1C may be related to fear memory recall in PTP.
In neuropathic pain, nociceptive transmission has been reported to involve Ca2+ influx from Cav1.2 LTCCs in dorsal horn neurons of the spinal cord. 18 The rs216009 SNP of the CACNA1C gene that encodes the α1C subunit of the Cav1.2 LTCC 3 is located in an intron region and isolated outside the LD block (Figure 1). CACNA1C transcription may be regulated by enhancer activity around the rs216009 SNP in the spinal trigeminal nucleus, although further studies are needed to confirm this possibility. The region around the rs216009 SNP may be involved in changes in CACNA1C expression as an enhancer of the cyclic adenosine monophosphate response element binding protein (CREB)-dependent promoter in the upstream region of the CACNA1C gene. 19 Ca2+ influx into cells through Cav1.2 channels has been shown to induce CREB activation, which depends on nociceptive activity.18,20 The rs216009 SNP may be involved in enhancing CACNA1C expression, thereby affecting the increase in Ca2+ influx and CREB activation. CREB activation, in turn, would lead to further CACNA1C expression 19 and the activation of pain-related genes. 21 The activation of pain-related genes may have been involved in the vulnerability to PTP in the present study through both an increase in intracellular Ca2+ levels by upregulated CACNA1C expression and CREB activation. Although the facts based on the previous reports and the present study seem to be logically consistent, further research is needed to confirm the mechanisms that underlie the vulnerability to PTP.
In the present study, C-allele homozygote carriers of the rs216009 SNP had a higher rate of PTP, suggesting that the C allele of the CACNA1C rs216009 SNP is associated with the vulnerability to PTP in an autosomal recessive manner. Allele frequencies of the rs216009 SNP of the CACNA1C gene in different regional populations and in the present study among patients with PTP and OFP and among total patients are as follows: The rs216009 SNP of the CACNA1C gene has a T-allele frequency of 62% and C-allele frequency of 38% in East Asian populations, according to the 1000 Genomes study in the dbSNP database. 22 The subjects in the present study were Japanese who had allele frequencies that were similar to the general East Asian population (total patients: 58% T allele, 42% C allele). Additionally, OFP patients had C-allele frequencies that were similar to East Asian populations (OFP patients: 63% T allele, 37% C allele). PTP patients had a T-allele frequency of 41% and C-allele frequency of 59%, with a higher percentage of C alleles compared with other regional populations (e.g., American populations: 78% T allele, 22% C allele; African populations: 84% T allele, 16% C allele; European populations: 90% T allele, 10% C allele; South Asian populations: 79% T allele, 21% C allele). In the present study, homozygous C-allele carriers of the rs216009 SNP had a higher incidence of PTP, suggesting that the C allele is associated with the susceptibility to PTP. These results suggest that Japanese and other East Asian populations, because of their higher C-allele frequency, may have a higher risk of PTP than populations in other regions.
In conclusion, homozygous carriers of the C-allele of the rs216009 SNP of the CACNA1C gene exhibited greater vulnerability to PTP, possibly through the regulation of intracellular Ca2+ levels and affective pain systems, such as those that mediate fear memory recall. Further research is needed to elucidate the precise mechanisms of PTP development.
Supplemental Material
Supplemental Material for The rs216009 single-nucleotide polymorphism of the CACNA1C gene is associated with phantom tooth pain by Masako Morii, Seii Ohka, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Moe Soeda, Ken-ichi Fukuda, Kaori Yoshida, Kyotaro Koshika, Tatsuya Ichinohe, and Kazutaka Ikeda in Molecular Pain
Acknowledgements
We thank Dr. Yoshiyuki Shibukawa for assistance with writing the manuscript and thought-provoking discussions and Mr. Michael Arends for assistance with editing the manuscript. We are grateful to the volunteers for their participation in the study and anesthesiologists and surgeons for collecting the clinical data.
Author’s Contribution: MM, SO, DN, KF, and KI conceived the study and designed the experiments. MM, SO, KN, YE, and DN performed the statistical analyses. MM, SO, and DN wrote the manuscript. MS and KF collected clinical samples and data. MM, MS, and JH performed the genotyping procedures. MM and SO performed the database analysis. SO, DN, MS, KF, KY, KK, TI, and KI supervised the experiments and finalized the manuscript. All authors contributed to writing the manuscript, and all authors read and approved the final manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the grants from the Japan Society for the Promotion of Science (JSPS) KAKENHI (no. 21H03028, JP22H04922 [AdAMS], 20K07774, 20K09259, 17K08970, 17H04324, and JP19K19212) and Japan Agency for Medical Research and Development (AMED; no. JP19ek0610011).
Supplemental Material: Supplemental material for this article is available online.
ORCID iDs
Seii Ohka https://orcid.org/0000-0003-2394-6440
Daisuke Nishizawa https://orcid.org/0000-0001-8286-4651
Kyotaro Koshika https://orcid.org/0000-0002-2656-9698
Kazutaka Ikeda https://orcid.org/0000-0001-8342-0278
References
- 1.Marbach JJ, Raphael KG. Phantom tooth pain: a new look at an old dilemma. Pain Med 2000; 1: 68–77. DOI: 10.1046/j.1526-4637.2000.00012.x. [DOI] [PubMed] [Google Scholar]
- 2.Soeda M, Ohka S, Nishizawa D, Hasegawa J, Nakayama K, Ebata Y, Fukuda K, Ikeda K. Single-nucleotide polymorphisms of the SLC17A9 and P2RY12 genes are significantly associated with phantom tooth pain. Mol Pain 2022; 18: 17448069221089592. DOI: 10.1177/17448069221089592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catterall WA, Perez-Reyes E, Snutch TP, Striessnig J. International Union of Pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol Rev 2005; 57: 411–425. DOI: 10.1124/pr.57.4.5. [DOI] [PubMed] [Google Scholar]
- 4.Catterall WA. Voltage-gated calcium channels. Cold Spring Harb Perspect Biol 2011; 3: a003947. DOI: 10.1101/cshperspect.a003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Q, Lu J, Zhou X, Chen X, Su D, Gu X, Yu W. High-voltage-activated calcium channel in the afferent pain pathway: an important target of pain therapies. Neurosci Bull 2019; 35: 1073–1084. DOI: 10.1007/s12264-019-00378-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radwani H, Lopez-Gonzalez MJ, Cattaert D, Roca-Lapirot O, Dobremez E, Bouali-Benazzouz R, Eiríksdóttir E, Ülo L, Favereaux A, Errami M, Landry M, Fossat P. Cav1.2 and Cav1.3 L-type calcium channels independently control short- and long-term sensitization to pain. J Physiol 2016; 594: 6607–6626, DOI: 10.1113/JP272725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu YP, Wu X, Xia X, Yao J, Wang BJ. The genome-wide supported CACNA1C gene polymorphisms and the risk of schizophrenia: an updated meta-analysis. BMC Med Genet 2020; 21: 159, DOI: 10.1186/s12881-020-01084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moon AL, Haan N, Wilkinson LS, Thomas KL, Hall J. CACNA1C: Association with psychiatric disorders, behavior, and neurogenesis. Schizophr Bull 2018; 44: 958–965. DOI: 10.1093/schbul/sby096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeon D, Kim S, Chetana M, Jo D, Ruley HE, Lin SY, Rabah D, Kinet JP, Shin HS. Observational fear learning involves affective pain system and Cav1.2 Ca2+ channels in ACC. Nat Neurosci 2010; 13: 482–488. DOI: 10.1038/nn.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.International classification of orofacial pain, 1st edition (ICOP). Cephalalgia 2020; 40: 129-221. DOI: 10.1177/0333102419893823 [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization portal . ICD-11 2022 release, WHO. https://icd.who.int/browse11/l-m/en. [Google Scholar]
- 12.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet 2005; 37: 1217–1223. DOI: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 13.GTEx portal on 09/22/2022 . Referred to gene page of CACNA1C, GTEX. https://www.gtexportal.org/. [Google Scholar]
- 14.Zhang Q, Manders T, Tong AP, Yang R, Garg A, Martinez E, Zhou H, Dale J, Goyal A, Urien L, Yang G, Chen Z, Wang J. Chronic pain induces generalized enhancement of aversion. eLife 2017; 6: e25302, DOI: 10.7554/eLife.25302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wager TD, Atlas LY, Lindquist MA, Roy M, Woo CW, Kross E. An fMRI-based neurologic signature of physical pain. N Engl J Med 2013; 368: 1388–1397, doi: 10.1056/NEJMoa1204471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bushnell MC, Ceko M, Low LA. Cognitive and emotional control of pain and its disruption in chronic pain. Nat Rev Neuroci 2013; 14: 502–511. DOI: 10.1038/nrn3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fossat P, Dobremez E, Bouali-Benazzouz R, Favereaux A, Bertrand S, Kilk K, Léger C, Cazalets JR, Langel Ü, Landry M, Nagy F. Knockdown of L calcium channel subtypes: differential effects in neuropathic pain. J Neurosci 2010; 30: 1073–1085. DOI: 10.1523/JNEUROSCI.3145-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bavley CC, Kabir ZD, Walsh AP, Kosovsky M, Hackett J, Sun H, Vázquez-Rosa E, Cintrón-Pérez CJ, Miller E, Koh Y, Pieper AA, Rajadhyaksha AM. Dopamine D1R-neuron cacna1c deficiency: a new model of extinction therapy-resistant post-traumatic stress. Mol Psychiatry 2021; 26: 2286–2298. DOI: 10.1038/s41380-020-0730-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roca-Lapirot O, Radwani H, Aby F, Nagy F, Landry M, Fossat P. Calcium signalling through L-type calcium channels: role in pathophysiology of spinal nociceptive transmission. Br J Clin Pharmacol 2018; 175: 2362–2374. DOI: 10.1111/bph.13747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai CT, Wang DL, Chen WP, Hwang JJ, Hsieh CS, Hsu KL, Tseng CD, Lai LP, Tseng YZ, Chiang FT, Lin JL. Angiotensin II increases expression of alpha1C subunit of L-type calcium channel through a reactive oxygen species and cAMP response element-binding protein-dependent pathway in HL-1 myocytes. Circ Res 2007; 25: 1476–1485. DOI: 10.1161/01.RES.0000268497.93085.e1. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y, Wen J, Zheng B, Wu S, Mao O, Liang L, Li Z, Bachmann T, Bekker A, Tao YX. CREB participates in paclitaxel-induced neuropathic pain genesis through transcriptional activation of Dnmt3a in primary sensory neurons. Neurotherapeutics 2021; 18: 586–600, DOI: 10.1007/s13311-020-00931-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.NCBI portal on 09/28/2022 . Referred to SNP report of rs216009 in dbSNP, NCBI. https://www.ncbi.nlm.nih.gov/snp/rs216009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material for The rs216009 single-nucleotide polymorphism of the CACNA1C gene is associated with phantom tooth pain by Masako Morii, Seii Ohka, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Moe Soeda, Ken-ichi Fukuda, Kaori Yoshida, Kyotaro Koshika, Tatsuya Ichinohe, and Kazutaka Ikeda in Molecular Pain