Abstract
Although numerous studies have suggested the association between TNF-α-308G/A polymorphism and susceptibility to obstructive sleep apnea (OSA), the results remained controversial and ambiguous. We performed the present meta-analysis to derive a more precise estimation.The PubMed, Embase, Cochrane library, Web of Science, China National Knowledge Infrastructure, Wanfang databases, and Weipu databases (until January 8, 2022) were accessed to retrieve relevant articles. Pooled odds ratios (ORs) with 95% confidence interval (95% CI) were calculated using the STATA statistical software.Totally, fourteen studies involving 2595 cases and 2579 controls were enrolled in this meta-analysis. Pooled results demonstrated significant association between TNF-α-308G/A polymorphism and OSA risk for the overall population(allele model:OR = 1.87 [1.47, 2.38] (n = 14), dominant model: OR = 1.88[1.48, 2.39] (n = 14), recessive model:OR = 2.83 [2.00, 4.00] (n = 11), homozygous model:OR = 3.30 [2.32, 4.68] (n = 11), and heterozygous model:OR = 1.67 [1.36, 2.06] (n = 14); P<0.001, respectively).Subgroup analysis showed that in both Caucasians and Asians, the A allele conferred increased risk to OSA compared to the G allele (Caucasians: OR = 1.40[1.03, 1.90] (n = 5), P = 0.033, Asians: OR = 2.30 [1.62, 3.26] (n = 9), P< 0.001). In subgroup analysis restricted to hospital-based individuals, significant association between TNF-α-308G/A polymorphism and OSA risk was identified under each genetic model. Whereas, in population-based individuals, increased risk of OSA were only found in homozygous model (OR = 2.19[1.23, 3.90] (n = 3), P = 0.008) and recessive model (OR = 1.77 [1.00, 3.13] (n = 3), P = 0.048). There was a substantial between-study heterogeneity (I2 = 69.10%) across studies which was explained by source of control participants (P = 0.036) by meta-regression. The results of leave-one-out meta-analysis and publication bias suggested the reliability and stability of our results.This meta-analysis suggested that TNF-α-308A allele may be a risk factor for the development of OSA. However, large scale,multi-center and well-designed case-control studies are needed in the future.
Introduction
Obstructive sleep apnea (OSA) is a prevalent sleep-related breathing disorder resulting from the repetitive obstruction of the upper airway during sleep with prevalence ranging from 9% to 38% in the general population. The prevalence rises with advancing age, male sex, and high body mass index [1]. Published studies reported the associations between OSA and increased risk of cardiovascular disease, cognitive impairment, cancer, and metabolic syndrome [2–5]. Although continuous positive airway pressure (CPAP) is the first-line therapy for OSA, improving its compliance and identifying novel therapy strategies are still actively studied. Thus, OSA remains a public health challenge around the world, up till now, its etiology and pathogenesis are not fully understood.
Recently, the heritability of OSA and the indices of OSA severity, such as apnea hypopnea index (AHI), respiratory disturbance index (RDI), and oxygen desaturation index (ODI) were reported by Szily and coworkers [6]. Moreover, published studies have shown that OSA is more prevalent in Asia than in Europe. Ethnic differences in severity and treatment were also illustrated, indicating the potential role of genetic factors in imparting the variance of OSA [7–9]. Genetic associations with OSA, including the tumor necrosis factor (TNF)-α-308G/A polymorphism, have been studied extensively [10]. This polymorphism is characterized by adenine (A) substituted for guanine (G), which induces the expression of TNF-α [11, 12]. Remarkably, compared with control subjects, patients with OSA have higher systemic TNF-α levels, which showed a positive correlation with OSA severity [13]. Taken together, these findings suggest that TNF-α-308G/A polymorphism may be involved in the pathogenesis of OSA.
Although numerous studies have investigated the association between TNF-α-308G/A polymorphism and susceptibility to OSA, the results remained controversial and ambiguous [14, 15], which may be related to ethnicity, source of control, genotyping method, sample size, and so on. TNF-α-308G/A polymorphism in the promoter region of TNF gene is reported to be closely associated with OSA risk [14, 16], whereas others are not [15, 17]. Despite previous meta-analyses have been conducted [18–21], a number of relevant studies are not enrolled in their meta-analyses [14–16, 22, 23]. Hence, we performed a meta-analysis to derive a more accurate evaluation of the association between TNF-α-308G/A polymorphism and OSA susceptibility in all available studies. Moreover, source of the control participants was firstly used for subgroup analyses and confirmed to be the potential source of between-study heterogeneity by meta-regression.
Materials and methods
Search strategy
The PubMed, Embase, Cochrane library, Web of Science, China National Knowledge Infrastructure, Wanfang databases, and Weipu databases (until January 8, 2022) were used to perform a systematic search for the identification of studies addressing the association of TNF-α-308 G/A polymorphism and OSA. The adopted appropriate combinations of search terms were as follows: “obstructive sleep apnea syndrome”, “OSAS”, “obstructive sleep apnea”, “OSA” and “tumor necrosis factor”, “TNF”, “tumor necrosis factor-a”, “TNF-α” and “polymorphism”, “gene”, “variant”, and “mutation”. A manual search was also performed by reviewing the references of relevant studies and review articles.
Inclusion and exclusion criteria
Studies that met the following criteria were included in the meta-analysis: (1) studies addressing the association of TNF-α-308 G/A polymorphism with OSA, (2) case–control design, (3) adequate genotype distributions in cases and controls, and (4) full-text studies published in English and Chinese. The exclusion criteria we adopted were as follows: (1) enrolled patients without OSA; (2) enrolled patients without available genotype distribution; (3) letter, editorial, review or meta-analysis, abstract, conference papers, case report, studies on animals or cell-lines; and (4) duplicate studies. The definition of OSA was as described in the primary studies.
Data extraction
The following data were extracted independently from each eligible study by two investigators (Zhang ZL and DD): first author’s surname, year of publication, country, ethnicity, age, body mass index, gender (male/female), AHI of cases and controls, source of control, controls matched for, genotype method, Hardy–Weinberg equilibrium (HWE) in controls, sample size, and genotype frequencies in cases and controls. Discrepancies were settled through consultation. We contacted the authors by email if detailed data were missing.
Quality assessment
The Newcastle–Ottawa Scale (NOS) captured the quality of eligible studies in terms of three parts, including “Selection”, “Comparability”, “Exposure”, stratified into by low- (0–3), moderate- (4–6), and high-quality (7–9) levels in accordance with scores.
Statistical analysis
The STATA statistical software (version 16.0) was used to carry out the meta-analyses. Pooled odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using the Z test to evaluate the association of TNF-α-308 G/A polymorphism with OSA risk under the following genetic models: dominant (AA + AG vs. GG), recessive (AA vs. AG + GG), homozygous (AA vs. GG), heterozygote (AG vs. GG) and allele (A allele vs. G allele) models. The Pearson’s χ2 test was performed to evaluate the HWE in control groups. The heterogeneity across studies was evaluated using the Cochran’s Q and the I2 statistics. Significant heterogeneity was considered when P < 0.1 for the Q-test or I2 > 50%. Thus, the random-effects model was applied to calculate the pooled ORs. Otherwise, the fixed-effects model was implemented. Subgroup analyses and meta-regression were conducted to explore potential sources of heterogeneity in total models according to ethnicity and source of control. The sensitivity analysis which was carried out with leave-one-out meta-analysis were performed for all genetic models to further confirm the reliability and stability of the results. Harbord funnel plot and Peters test were performed to access the publication bias for all models. P-value < 0.05 was considered to indicate statistical significance.
Results
Selection of studies
The process of study selection was presented in Fig 1. A total of 437 studies were initially identified through the database described above. 196 studies were screened after removing duplicates. By screening titles and abstracts, 148 studies were excluded because they were 1) meta-analyses or reviews (n = 29), 2) letters, editorials, abstracts, and case reports (n = 10), 3) studies on animals or cell lines (n = 61), 4) studies not on TNF-α (n = 34), studies not for case-control study(n = 1), and 5) studies not on OSA (n = 13). A total of 48 full-text articles were further assessed for eligibility, and 34 articles were excluded because they were not for TNF-α rs1800629 (n = 33) or had insufficient data (n = 1) [24]. Finally, 14 studies were eligible for the inclusion criteria and enrolled in this meta-analysis with a total number of 5174 subjects consisting of 2595 cases and 2579 controls [14–17, 22, 23, 25–32].
Fig 1. Flow diagram of study selection (PRISMA format).
Characteristics of the included studies
As summarized in Table 1, all eligible studies were published from 2005 to 2021. In terms of study populations, nine studies were conducted in Asian and five in Caucasian. The sources of the control participants were hospital-based (HB) in nine studies and population-based (PB) in five studies. The polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP), TaqMan assay, DNA sequencing, KASP and Mass Array technology were used to identify the genotype in eligible studies. The HWE test was conducted for the control group in each eligible study. Only one study did not conform to the HWE law (P < 0.05) [26]. All eligible studies were of moderate or high quality according to the NOS quality assessment.
Table 1. Basic characteristics of included studies.
| author | year | case | control | case | control | HWE | source of control participants | genotype method | country | ethnicity | Quality score | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | AG | AA | GG | AG | AA | ||||||||||
| Riha [32] | 2005 | 103 | 190 | 52 | 44 | 7 | 130 | 52 | 8 | 0.344 | PB | TaqMan | UK | Caucasian | 8 |
| Li [30] | 2006 | 24 | 48 | 11 | 10 | 3 | 35 | 12 | 1 | 0.981 | HB | TaqMan | China | Asian | 8 |
| Liu [31] | 2006 | 76 | 42 | 45 | 23 | 8 | 35 | 6 | 1 | 0.268 | HB | PCR-RFLP | China | Asian | 8 |
| Popko [28] | 2008 | 102 | 77 | 73 | 29 | 0 | 59 | 18 | 0 | 0.246 | PB | PCR-RFLP | Poland | Caucasian | 9 |
| Bhushan [26] | 2009 | 104 | 103 | 74 | 22 | 8 | 90 | 10 | 3 | 0.001 | HB | PCR-RFLP | Indians | Asian | 8 |
| Karkucak [27] | 2012 | 69 | 42 | 51 | 18 | 0 | 33 | 9 | 0 | 0.437 | HB | PCR-RFLP | Turkish | Caucasian | 8 |
| Almpanidou [25] | 2012 | 220 | 319 | 118 | 83 | 19 | 221 | 84 | 14 | 0.107 | PB | PCR-RFLP | Greek | Caucasian | 9 |
| Guan [29] | 2013 | 531 | 162 | 430 | 95 | 6 | 143 | 18 | 1 | 0.604 | HB | PCR-RFLP | China | Asian | 8 |
| Li [17] | 2013 | 155 | 100 | 137 | 18 | 0 | 88 | 12 | 0 | 0.523 | PB | DNA sequencing | China | Asian | 8 |
| Wang [22] | 2014 | 78 | 78 | 54 | 16 | 8 | 66 | 10 | 2 | 0.057 | HB | PCR-RFLP | China | Asian | 8 |
| Petrek [15] | 2015 | 116 | 351 | 85 | 30 | 1 | 248 | 98 | 5 | 0.175 | PB | Mass Array technology | Czech | Caucasian | 5 |
| Zhu [16] | 2015 | 200 | 200 | 124 | 46 | 30 | 166 | 31 | 3 | 0.278 | HB | DNA sequencing | China | Asian | 8 |
| Zhang [14] | 2019 | 750 | 800 | 576 | 153 | 21 | 656 | 132 | 12 | 0.077 | HB | TaqMan | China | Asian | 8 |
| Abulikemu [23] | 2021 | 67 | 67 | 42 | 20 | 5 | 60 | 6 | 1 | 0.098 | HB | KASP | China | Asian | 7 |
Abbreviation
HWE, Hardy-Weinberg equilibrium
PB,population-based
HB,hospital-based
PCR-RFLP:polymerase chain reaction-restriction fragment length polymorphism
KASP,kompetitive allele specific PCR.
Quantitative synthesis
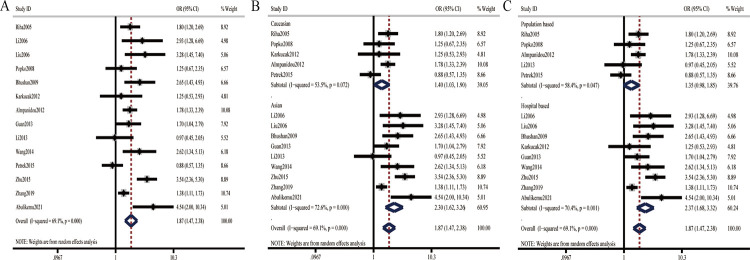
Pooled results demonstrated significant association between TNF-α-308G/A polymorphism and OSA risk for overall population under specific genetic models (Fig 2A and Table 2) (allele model:OR = 1.87 [1.47, 2.38] (n = 14), dominant model: OR = 1.88[1.48, 2.39] (n = 14), recessive model:OR = 2.83 [2.00, 4.00] (n = 11), homozygous model:OR = 3.30 [2.32, 4.68] (n = 11), and heterozygous model:OR = 1.67[1.36, 2.06] (n = 14); P<0.001, respectively).Subgroup analysis according to ethnicity showed that in both Caucasians and Asians, the A allele conferred increased risk to OSA compared to the G allele(Fig 2B and Table 2) (Caucasians: OR = 1.40[1.03, 1.90] (n = 5), P = 0.033, Asians: OR = 2.30 [1.62, 3.26] (n = 9), P< 0.001). In subgroup analysis restricted to HB individuals, significant association between TNF-α-308G/A polymorphism and OSA risk was identified under each genetic model (Fig 2C and Table 2). Whereas, in PB individuals, increased risk of OSA were only found in homozygous model (OR = 2.19[1.23, 3.90] (n = 3), P = 0.008) and recessive model (OR = 1.77 [1.00, 3.13] (n = 3), P = 0.048) (Table 2).
Fig 2. Forest plots of the association between TNF-α-308G/A polymorphism and OSA under the allele model.
A: overall results; B: subgroup analysis by ethnicity; C: subgroup analysis by source of control.
Table 2. Meta-analysis results of association between TNF-α-308G/A polymorphism and OSA.
| Contrast model | Subjects (cases/controls) | Studies,n | OR (95%CI) | P * | I2(%) | |
|---|---|---|---|---|---|---|
| Total studies | ||||||
| allele model | A vs. G | 5190/5158 | 14 | 1.87 (1.47, 2.38) | 0 | 69.10 |
| dominant model | AA/AG vs. GG | 2595/2579 | 14 | 1.88(1.48, 2.39) | 0 | 59.50 |
| recessive model | AA vs. AG/GG | 2269/2360 | 11 | 2.83 (2.00, 4.00) | 0 | 15.00 |
| homozygous model | AA vs. GG | 2269/2360 | 11 | 3.30 (2.32, 4.68) | 0 | 20.80 |
| heterozygous model | AG vs. GG | 2595/2579 | 14 | 1.67 (1.36, 2.06) | 0 | 38.80 |
| Subgroup analysis | ||||||
| Caucasians | ||||||
| allele model | A vs. G | 1220/1958 | 5 | 1.40(1.03, 1.90) | 0.033 | 53.50 |
| dominant model | AA/AG vs. GG | 610/979 | 5 | 1.48 (1.03, 2.13) | 0.035 | 56.30 |
| recessive model | AA vs. AG/GG | 439/860 | 3 | 1.77 (1.00, 3.13) | 0.048 | 0.00 |
| homozygous model | AA vs. GG | 439/860 | 3 | 2.19 (1.23, 3.90) | 0.008 | 0.00 |
| heterozygous model | AG vs. GG | 610/979 | 5 | 1.46 (1.04, 2.06) | 0.029 | 48.10 |
| Asians | ||||||
| allele model | A vs. G | 3970/3200 | 9 | 2.30 (1.62, 3.26) | 0 | 72.60 |
| dominant model | AA/AG vs. GG | 1985/1600 | 9 | 2.23 (1.59, 3.14) | 0 | 62.80 |
| recessive model | AA vs. AG/GG | 1830/1500 | 8 | 3.51 (2.06, 5.98) | 0 | 11.00 |
| homozygous model | AA vs. GG | 1830/1500 | 8 | 4.22 (2.32, 7.67) | 0 | 23.10 |
| heterozygous model | AG vs. GG | 1985/1600 | 9 | 1.86 (1.40, 2.47) | 0 | 39.60 |
| Subgroup analysis | ||||||
| HB | ||||||
| allele model | A vs. G | 3798/3084 | 9 | 2.37 (1.68, 3.32) | 0 | 70.40 |
| dominant model | AA/AG vs. GG | 1899/1542 | 9 | 2.30 (1.66, 3.20) | 0 | 58.70 |
| recessive model | AA vs. AG/GG | 1830/1500 | 8 | 3.51 (2.06, 5.98) | 0 | 11.00 |
| homozygous model | AA vs. GG | 1830/1500 | 8 | 4.22 (2.32, 7.67) | 0 | 23.10 |
| heterozygous model | AG vs. GG | 1899/1542 | 9 | 1.89 (1.45, 2.47) | 0 | 31.40 |
| PB | ||||||
| allele model | A vs. G | 1392/2074 | 5 | 1.35 (0.98, 1.85) | 0.068 | 58.40 |
| dominant model | AA/AG vs. GG | 696/1037 | 5 | 1.41 (0.97, 2.06) | 0.074 | 61.40 |
| recessive model | AA vs. AG/GG | 439/860 | 3 | 1.77 (1.00, 3.13) | 0.048 | 0.00 |
| homozygous model | AA vs. GG | 439/860 | 3 | 2.19 (1.23, 3.90) | 0.008 | 0.00 |
| heterozygous model | AG vs. GG | 696/1037 | 5 | 1.40 (0.98, 2.00) | 0.066 | 54.50 |
Abbreviation
HB: hospital-based
PB: population-based
Note:P*:P value for pooled OR.
Heterogeneity and sensitivity analysis
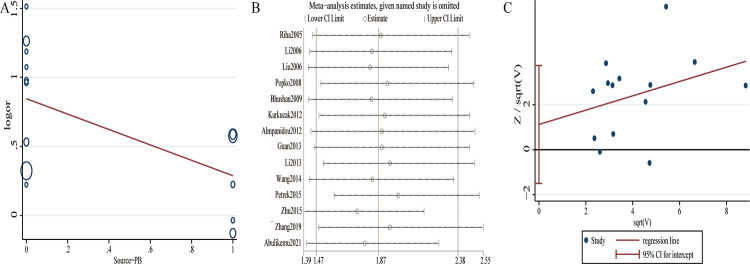
Significant between-study heterogeneity was considered in overall pooled analysis under allele, dominant and heterozygous models. Thus, random-effects model was applied to calculate the pooled ORs. Meta-regression was conducted to explore potential source of between-study heterogeneity, showing that the source of control participants caused the heterogeneity between studies in allele model (P = 0.036) (Fig 3A). However, we did not find the potential source of heterogeneity in dominant(P = 0.216 for ethnicity, P = 0.124 for source of control participants) and heterozygous models(P = 0.303 for ethnicity, P = 0.181 for source of control participants). Additionally, as presented in Fig 3B, results of leave-one-out meta-analysis showed no significant effect on the pooled ORs by any single study. Moreover, while omitting one study without confirmation of HWE [26], pooled ORs were not noticeably changed (OR = 1.83[1.42, 2.35]), further confirming the reliability and stability of present meta-analysis.
Fig 3. Meta-regression, sensitivity analysis and publication bias in overall population under the allele model.
A: meta-regression(P = 0.036); B: sensitivity analysis; C: Harbord funnel plot (Harbord test P = 0.368, Peters test P = 0.120).
Publication bias
Harbord funnel plot and Peters test were performed to estimate publication bias under all genetic models, and no evidence of publication bias was found in this meta-analysis (A vs. G: Harbord test P = 0.368, Peters test P = 0.120; AA/AG vs. GG: Harbord test P = 0.212, Peters test P = 0.097; AA vs. AG/GG: Harbord test P = 0.810, Peters test P = 0.300; AA vs. GG: Harbord test P = 0.921, Peters test P = 0.266; AG vs. GG: Harbord test P = 0.131, Peters test P = 0.084) (Fig 3C).
Discussion
On the basis of 14 studies involving 2595 cases and 2579 controls, the present meta-analysis aimed to clarify the association between TNF-α-308G/A polymorphism and susceptibility of OSA. Pooled results demonstrated a significant association of TNF-α-308A allele and increased risk of OSA (OR = 1.87 [1.47, 2.38], P < 0.001), indicating that TNF-α-308A allele may be a risk factor for the development of OSA.
Furthermore, results of subgroup analysis remarkably showed a 1.40-fold increased risk of OSA for Caucasians and a 2.30-fold increased risk of OSA for Asians with allele A, suggesting the significant influence of ethnicity on OSA risk. This finding is consistent under different genetic models and may explain the higher prevalence of OSA in Asians than in Europeans [9]. Moreover, under allele model, stratified analysis by source of control showed increased risk of OSA in HB studies (OR = 2.37 [1.68, 3.32], P < 0.001) but not in PB studies (P = 0.068), indicating that certain aspects of the study design may affect the results of a case–control study. Besides, results of the codominant model proved that carrying either genotype AA or genotype AG would put people at higher risk for OSA, which is consistent with the findings that level of TNF-α for individuals with genotypes AG and AA is higher than that of individuals with genotype GG [33].
Evidence showed that TNF inhibition was associated with decreased risk of OSA [34]. Interestingly, evidence demonstrated a high level of circulating TNF-α in patients with OSA, indicating that TNF-α may be a promising circulating biomarker to assess the degree of OSA [13, 35–37]. We have comprehensively analyzed the results of 14 studies through present meta-analysis and found increased risk of OSA in those with TNF-α-308A allele, which may be explained by elevated TNF-α expression via regulating the promoter transcriptional activity [12].Moreover,TNF-α gene lies in the highly polymorphic major histocompatibility complex (MHC) region. Apart from TNF-α-308 G/A polymorphism, TNF-α-238 G/A (rs361525), TNF-α-857G/A (rs1799724), and TNF-α-1031 T/C (rs1799964) are identified. Therefore, the linkage disequilibrium between alleles across the MHC may be involved in regulating the expression of TNF-α. In addition to gene-gene interactions, gene-environment interactions may also play a role in regulating TNF-α expression and thus participating in the pathogenesis of OSA.Thus, although our results showed that TNF-α-308G/A polymorphism was an important gene locus affecting risk of OSA, the mechanism is complex and should be further studied.
Previous meta-analyses were conducted by Varvarigou et al. in 2011 (three studies) [20], Huang et al. in 2012 (four studies) [21], Zhong et al. in 2014 (seven studies) [19], and Wu et al. in 2014 (ten studies) [18]. Similar to the above meta-analyses, our results demonstrated a significant association of TNF-α-308A allele and increased risk of OSA. However, our meta-analysis was on the basis of 14 case–control studies involving 2595 cases,while the largest sample size in previous meta-analyses was 10 studies with 1522 patients [18], providing more convincing and precise evidence that TNF-α-308G/A polymorphism increased the susceptibility to OSA. Statistical similar OSA risk were observed both in European (1.68-fold) and Asian population (2.02-fold) carrying A allele by Wu et al. [18], whereas higher risk of OSA for Asians with allele A(2.30-fold) than Caucasians(1.40-fold) was found in the present meta-analysis. Moreover, source of control participants was firstly used for subgroup analyses, showing that TNF-α-308A allele increases the risk of OSA in HB case–control studies but not in PB studies, suggesting the influence of study design on results. Additionally, we further performed meta-regression and found that source of control participants may be the potential source of heterogeneity
Study limitations
Although the results of sensitivity analysis and publication bias suggested the stability of our results, some limitations should be considered. First, language bias resulted from limitation to publication in English and Chinese. Second, the confounding bias created by unavailable factors in most eligible studies like sex, age, body mass index, life style, AHI, comorbidities, other gene polymorphisms that may play important roles in the pathophysiology of OSA. Third, sampling bias resulted from enrolling a study unconforming to HWE law [26]. Fourth, there could be additional factors that have not been included in this review.
In summary, the present meta-analysis indicated significant association between allele A and increased susceptibility of OSA. However, large scale,multi-center and well-designed case-control studies are also needed in the future. Meanwhile, it is also of great significance to explore the mechanism of TNF-α-308 G/A polymorphism involved in OSA pathogenesis.
Supporting information
(DOCX)
(DOCX)
A: overall results; B: subgroup analysis by ethnicity; C: subgroup analysis by source of control.
(TIF)
A: overall results; B: subgroup analysis by ethnicity; C: subgroup analysis by source of control.
(TIF)
A: overall results; B: subgroup analysis by ethnicity; C: subgroup analysis by source of control.
(TIF)
A: overall results; B: subgroup analysis by ethnicity; C: subgroup analysis by source of control.
(TIF)
Acknowledgments
We appreciate the contribution of participants and researchers in the studies included in this review.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by Natural Science Foundation of Xinjiang Uygur Autonomous Region (No.2021D01C278) received by ZZL. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Senaratna CV, Perret JL, Lodge CJ, Lowe AJ, Campbell BE, Matheson MC, et al. Prevalence of obstructive sleep apnea in the general population: A systematic review. Sleep Med Rev. 2017;34:70–81. doi: 10.1016/j.smrv.2016.07.002 [DOI] [PubMed] [Google Scholar]
- 2.Gabryelska A, Karuga FF, Szmyd B, Bialasiewicz P. HIF-1alpha as a Mediator of Insulin Resistance, T2DM, and Its Complications: Potential Links With Obstructive Sleep Apnea. Front Physiol. 2020;11:1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mullins AE, Kam K, Parekh A, Bubu OM, Osorio RS, Varga AW. Obstructive Sleep Apnea and Its Treatment in Aging: Effects on Alzheimer’s disease Biomarkers, Cognition, Brain Structure and Neurophysiology. Neurobiol Dis. 2020;145:105054. doi: 10.1016/j.nbd.2020.105054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Z, Wang L, Liu Y, Li G, Chen J. Obstructive sleep apnea in coronary artery disease: the role of nocturnal hypoxic burden. J Clin Sleep Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gozal D, Almendros I, Phipps AI, Campos-Rodriguez F, Martinez-Garcia MA, Farre R. Sleep Apnoea Adverse Effects on Cancer: True, False, or Too Many Confounders? Int J Mol Sci. 2020;21(22). doi: 10.3390/ijms21228779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szily M, Tarnoki AD, Tarnoki DL, Kovacs DT, Forgo B, Lee J, et al. Genetic influences on the onset of obstructive sleep apnoea and daytime sleepiness: a twin study. Respir Res. 2019;20(1):125. doi: 10.1186/s12931-019-1095-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andreozzi F, Van Overstraeten C, Ben YS, Bold I, Carlier S, Gruwez A, et al. African ethnicity is associated with a higher prevalence of diabetes in obstructive sleep apnea patients: results of a retrospective analysis. Sleep Breath. 2020;24(3):857–64. doi: 10.1007/s11325-019-01912-5 [DOI] [PubMed] [Google Scholar]
- 8.Dudley KA, Patel SR. Disparities and genetic risk factors in obstructive sleep apnea. Sleep Med. 2016;18:96–102. doi: 10.1016/j.sleep.2015.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hnin K, Mukherjee S, Antic NA, Catcheside P, Chai-Coetzer CL, McEvoy D, et al. The impact of ethnicity on the prevalence and severity of obstructive sleep apnea. Sleep Med Rev. 2018;41:78–86. doi: 10.1016/j.smrv.2018.01.003 [DOI] [PubMed] [Google Scholar]
- 10.Mukherjee S, Saxena R, Palmer LJ. The genetics of obstructive sleep apnoea. Respirology. 2018;23(1):18–27. doi: 10.1111/resp.13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci U S A. 1997;94(7):3195–9. doi: 10.1073/pnas.94.7.3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Louis E, Franchimont D, Piron A, Gevaert Y, Schaaf-Lafontaine N, Roland S, et al. Tumour necrosis factor (TNF) gene polymorphism influences TNF-alpha production in lipopolysaccharide (LPS)-stimulated whole blood cell culture in healthy humans. Clin Exp Immunol. 1998;113(3):401–6. doi: 10.1046/j.1365-2249.1998.00662.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, Zheng X. Tumor necrosis factor alpha is a promising circulating biomarker for the development of obstructive sleep apnea syndrome: a meta-analysis. Oncotarget. 2017;8(16):27616–26. doi: 10.18632/oncotarget.15203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z, Wang Q, Chen B, Wang Y, Miao Y, Han L. Association study of genetic variations of inflammatory biomarkers with susceptibility and severity of obstructive sleep apnea. Mol Genet Genomic Med. 2019;7(8):e801. doi: 10.1002/mgg3.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrek M, Petrkova J, Zizkova V, Kolek V, Hobzova M. The rs1800629 variant in the TNF-alpha gene is not associated with obstructive sleep apnoea syndrome in a Czech population. EUROPEAN RESPIRATORY JOURNAL. 2015;46. [Google Scholar]
- 16.Zhu LL, Mai JY. Association of TNF alpha—308 loci gene polymorphism with risk of obstructive sleep apnea syndrome. Shandong Medical Journal. 2015. (36):53–4. [Google Scholar]
- 17.Li TZ, Qian XS, Guo RB, Sun BJ, Liu CC. Relation between TNF-α polymorphism and obstructive sleep apnea hypopnea syndrome accompanying hypertension. Acad J Chin PLA Med Sch. 2013. (12):1265–8. [Google Scholar]
- 18.Wu Y, Cao C, Wu Y, Zhang C, Zhu C, Ying S, et al. TNF-alpha-308G/A polymorphism contributes to obstructive sleep apnea syndrome risk: evidence based on 10 case-control studies. PLoS One. 2014;9(9):e106183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong A, Xiong X, Xu H, Shi M. An updated meta-analysis of the association between tumor necrosis factor-alpha -308G/A polymorphism and obstructive sleep apnea-hypopnea syndrome. PLoS One. 2014;9(9):e106270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varvarigou V, Dahabreh IJ, Malhotra A, Kales SN. A review of genetic association studies of obstructive sleep apnea: field synopsis and meta-analysis. Sleep. 2011;34(11):1461–8. doi: 10.5665/sleep.1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, Liao N, Huang QP, Xie ZF. Association between tumor necrosis factor-alpha-308G/A polymorphism and obstructive sleep apnea: a meta-analysis. Genet Test Mol Biomarkers. 2012;16(4):246–51. [DOI] [PubMed] [Google Scholar]
- 22.Wang HT, Ling M. Relationship between polymorphism of tumor necrosis factor-a gene rs1800629 and obstructive sleep apnea-hypopnea syndrome in Uygur population. J Chin Pract Diagn Ther,Dec. 2014;28(12):1172–4. [Google Scholar]
- 23.Abulikemu Y, Abulajang T, Ailigen A, Tang L. [Analysis of propensity score matching between inflammatory factor levels and gene polymorphisms and susceptibility to obstructive sleep apnea]. Lin chuang er bi yan hou tou jing wai ke za zhi = Journal of clinical otorhinolaryngology, head, and neck surgery. 2021. Aug;35(8):728–32. doi: 10.13201/j.issn.2096-7993.2021.08.012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khalyfa A, Serpero LD, Kheirandish-Gozal L, Capdevila OS, Gozal D. TNF-alpha Gene Polymorphisms and Excessive Daytime Sleepiness in Pediatric Obstructive Sleep Apnea. JOURNAL OF PEDIATRICS. 2011;158(1):81–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almpanidou P, Hadjigeorgiou G, Gourgoulianis K, Papadimitriou A. Association of tumor necrosis factor-alpha gene polymorphism (-308) and obstructive sleep apnea-hypopnea syndrome. HIPPOKRATIA. 2012;16(3):217–20. [PMC free article] [PubMed] [Google Scholar]
- 26.Bhushan B, Guleria R, Misra A, Luthra K, Vikram NK. TNF-alpha gene polymorphism and TNF-alpha levels in obese Asian Indians with obstructive sleep apnea. Respiratory medicine. 2009;103(3):386–92. doi: 10.1016/j.rmed.2008.10.001 [DOI] [PubMed] [Google Scholar]
- 27.Karkucak M, Ursavaş A, Ocakoǧlu G, Görükmez O, Yakut T, Ercan I, et al. Analysis of TNF-alpha G308A and C857T gene polymorphisms in Turkish patients with obstructive sleep apnea syndrome. Turkiye Klinikleri Journal of Medical Sciences. 2012;32(5):1368–73. [Google Scholar]
- 28.Popko K, Gorska E, Potapinska O, Wasik M, Stoklosa A, Plywaczewski R, et al. FREQUENCY OF DISTRIBUTION OF INFLAMMATORY CYTOKINES IL-1, IL-6 AND TNF-alpha GENE POLYMORPHISM IN PATIENTS WITH OBSTRUCTIVE SLEEP APNEA. Journal of physiology and pharmacology: an official journal of the Polish. 2008;59 Suppl 6:607–14. [PubMed] [Google Scholar]
- 29.Guan J, Yi HL, Meng LL, Su KM, Yin SK. Study of association of TNF-alpha gene Polymorphism with OSAHS and metabolic syndrome. CHlN ARCH OTOLARYNGOL HEAD NECK SURG. 2013;20(01):8–12. [Google Scholar]
- 30.Li FY. Tumor necrosis factor-a(-308)gene and serotonin transporter gene polymorphism with obstructive apnea syndrome: Shandong university, M1—master; 2006. [Google Scholar]
- 31.Liu HG, Guan P, Lin M, Xu YJ, Zhang ZX. The relationship between tumor necrosis factor-a gene promoter polymorphism and obstructive sleep apnea-hypopnea syndrome. Chin J TUberc Respir Dis. 2006;29(9):596–9. [PubMed] [Google Scholar]
- 32.Riha RL, Brander P, Vennelle M, McArdle N, Kerr SM, Anderson NH, et al. Tumour necrosis factor-alpha (-308) gene polymorphism in obstructive sleep apnoea-hypopnoea syndrome. The European respiratory journal. 2005. Oct;26(4):673–8. doi: 10.1183/09031936.05.00130804 . [DOI] [PubMed] [Google Scholar]
- 33.Peng Y, Li LJ. TNF-alpha-308G/A polymorphism associated with TNF-alpha protein expression in patients with diabetic nephropathy. Int J Clin Exp Pathol. 2015;8(3):3127–31. [PMC free article] [PubMed] [Google Scholar]
- 34.Walsh JA, Duffin KC, Crim J, Clegg DO. Lower frequency of obstructive sleep apnea in spondyloarthritis patients taking TNF-inhibitors. J Clin Sleep Med. 2012;8(6):643–8. doi: 10.5664/jcsm.2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao Y, Song Y, Ning P, Zhang L, Wu S, Quan J, et al. Association between tumor necrosis factor alpha and obstructive sleep apnea in adults: a meta-analysis update. BMC Pulm Med. 2020;20(1):215. doi: 10.1186/s12890-020-01253-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nadeem R, Molnar J, Madbouly EM, Nida M, Aggarwal S, Sajid H, et al. Serum inflammatory markers in obstructive sleep apnea: a meta-analysis. J Clin Sleep Med. 2013;9(10):1003–12. doi: 10.5664/jcsm.3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wali SO, Al-Mughales J, Alhejaili F, Manzar MD, Alsallum F, Almojaddidi H, et al. The utility of proinflammatory markers in patients with obstructive sleep apnea. Sleep Breath. 2020. doi: 10.1007/s11325-020-02149-3 [DOI] [PubMed] [Google Scholar]