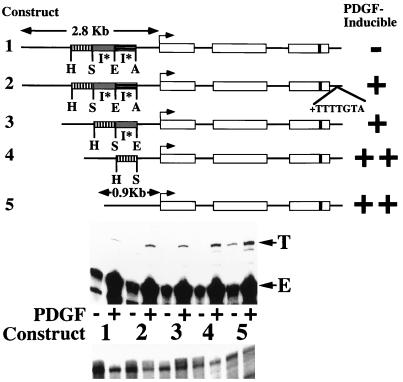
FIG. 1.
Deletion of 5′ flanking sequences results in heptamerless PDGF-inducible MCP-1 reporter genes. At the top is a schematic of the structures of five tagged MCP-1 reporter constructs. Open rectangles represent exons, and introns are represented by the dark lines between the exons. The lengths of 5′ flanking MCP-1 sequences contained within constructs are given in kilobases. The start of transcription is shown by the bent arrow. The 33-bp tag is represented by the dark band in the third exon. Constructs 1, 3, 4, and 5 contain 104 bp of 3′ untranslated sequences (that include the polyadenylation signal but not the heptamer). Only construct 2 contains the heptamer within its 3′ untranslated sequences. The relative positions of AocI (A), EcoRI (E), SpeI (S), and HincII (H) sites, located in the MCP-1 5′ flanking sequences and defining the endpoints of three MCP-1 5′ sequence fragments, are shown. The overall AocI-HincII 5′ fragment includes 1,936 bp. Construct 2 is derived from construct 1 by readdition of the heptamer TTTTGTA to the 3′ untranslated sequences of construct 1. Construct 3 is derived from construct 1 by deletion of the 707-bp EcoRI-AocI fragment. Construct 4 is derived from construct 3 by deletion of the 695-bp SpeI-EcoRI fragment. Construct 5 is derived from construct 4 by deletion of the 534-bp HincII-SpeI fragment. The PDGF-regulated 5′ I* elements are indicated. The PDGF inducibilities of the five constructs in transfection experiments are summarized on the right. In the middle are RNase protection assays of 40 μg of total cellular RNA that was prepared from NIH 3T3 fibroblasts transiently transfected with 5 μg of the constructs shown, allowed to become quiescent, and then not exposed (−) or exposed (+) to the B-B isoform of PDGF (30 ng/ml) for 3 h. The numbers refer to the tagged MCP-1 constructs diagrammed at the top. The positions of the 305- and 241-nt protected fragments corresponding to expression of the transfected and tagged (T) and endogenous (E) MCP-1 genes, respectively, are shown. The experiment was performed four times with similar results. The PDGF inductions obtained with constructs 4 and 5 were 2.6 to 4.0 and 3.0 to 8.1 times greater, respectively, than those obtained with construct 1 in these transfections. The PDGF induction increases observed with constructs 2, 3, 4, and 5, compared to construct 1, are all significant (P < 0.05 by the Wilcoxon two-sample test). At the bottom are RNase protection assays of 15 μg of total cellular RNA taken from the transfections shown above and analyzed with an alpha-globin riboprobe.