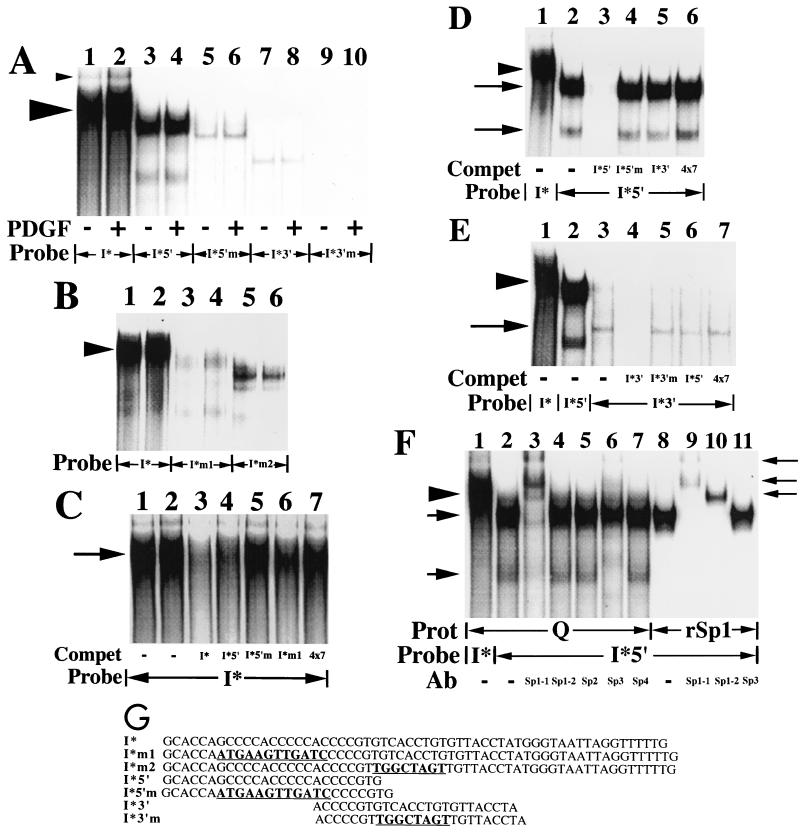
FIG. 6.
A slowly migrating protein complex binds to the 59-nt I* element-containing sequence. (A) Nuclear extracts (20 μg) prepared from quiescent fibroblasts (−) or fibroblasts treated with the B-B isoform of PDGF (30 ng/ml) for 2 h (+) were used in mobility shift assays along with the radiolabeled double-stranded oligonucleotide probes shown. The large arrowhead highlights the predominant complex that binds to the 59-nt fragment (I*) probe. The small arrowhead highlights an additional protein complex, of unclear significance, that also binds to the 59-nt fragment probe. (B) Nuclear extracts (15 μg) prepared from quiescent fibroblasts were used in mobility shift assays along with the radiolabeled double-stranded oligonucleotide probes shown. The arrowhead highlights the predominant complex that binds to the I* probe. (C) Nuclear extracts (15 μg) prepared from quiescent fibroblasts were used in mobility shift assays along with the radiolabeled double-stranded oligonucleotide probe shown. Unlabeled double-stranded oligonucleotides (300-fold excesses) were used as competitors where indicated. The 4×7 competitor is a double-stranded oligonucleotide containing four copies of the heptamer TTTTGTA. The arrow highlights the predominant complex that binds to the I* probe. Compet, competitor. (D) Nuclear extracts (15 μg) prepared from quiescent fibroblasts were used in mobility shift assays along with the radiolabeled double-stranded oligonucleotide probes shown. Unlabeled double-stranded oligonucleotides (300-fold excesses) were used as competitors where indicated. The arrowhead highlights the predominant complex that binds to the 59-nt fragment (I*) probe. The upper and lower arrows show the positions of two complexes that bind specifically to the I*5′ probe. (E) Nuclear extracts (20 μg) prepared from quiescent fibroblasts were used in mobility shift assays along with the radiolabeled double-stranded oligonucleotide probes shown. Unlabeled double-stranded oligonucleotides (200-fold excesses) were used as competitors where indicated. The arrowhead highlights the predominant complex that binds to the 59-nt fragment (I*) probe. The arrow shows the position of a complex that binds specifically to the I*3′ probe. (F) Nuclear extracts (15 μg) prepared from quiescent fibroblasts (Q) and recombinant Sp1 (rSp1, 1fpu) were used in mobility shift assays along with the radiolabeled double-stranded oligonucleotide probes shown. Antibodies specific for individual members of the Sp1 family of transcription factors (5 μg) were added where indicated. The Sp1-1 and Sp1-2 antibodies bind to distinct, nonoverlapping portions of the Sp1 transcription factor (amino acids 436 to 454 and 520 to 538, respectively). Prot, protein. The arrowhead on the left highlights the predominant complex that binds specifically to the 59-nt fragment (I*) probe. The arrows on the left show the positions of the two complexes that bind specifically to the 5′ subregion (I*5′) probe. The arrows on the right show the positions of complexes supershifted by the anti-Sp1 and anti-Sp3 antibodies (Ab). (G) The sequences of seven oligonucleotides and the relative position of each oligonucleotide within the overall 59-nt footprinted (I*) region are shown. These oligonucleotides were used as double-stranded probes or competitors in the six mobility shift assays whose results are shown here and correspond to the 25-nt (5′ subregion), 24-nt (3′ subregion), and 59-nt (full-length) footprinted regions shown in Fig. 3 (designated I*5′, I*3′, and I*, respectively). Mutant 25- and 24-nt probes, containing the 12- and 8-nt mutations described in the legend to Fig. 4 are designated I*5′m and I*3′m, respectively. Mutant I* probes containing the same 12- and 8-nt mutations are designated I*m1 and I*m2, respectively. The mutated sequences are boldfaced and underlined for each oligonucleotide. Free probe is not shown in these mobility shift assays. No complexes were observed with any of the probes alone in the absence of extract (data not shown).