Fig. 1.
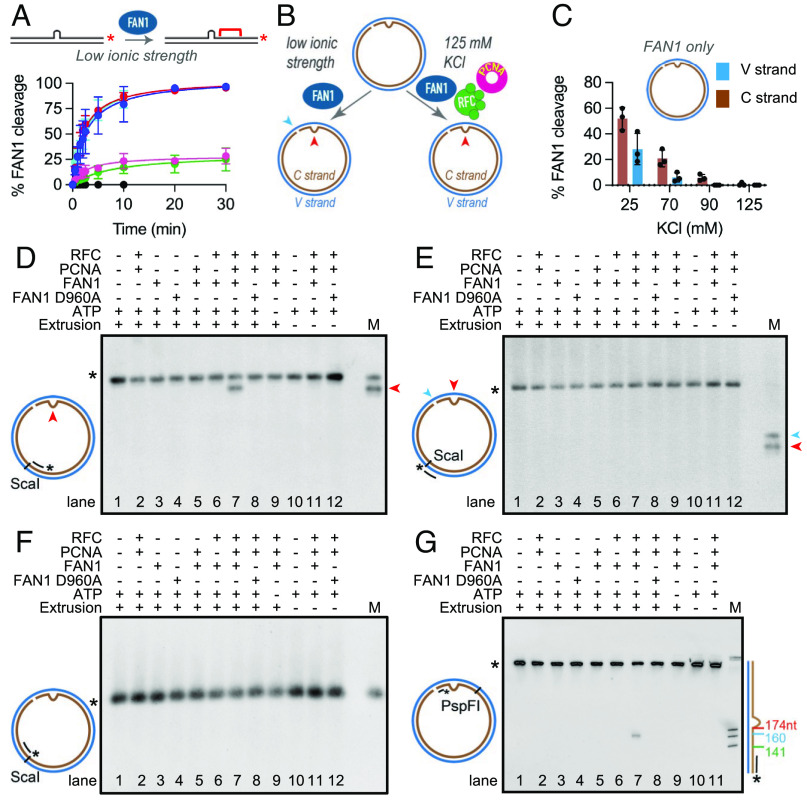
PCNA and RFC activate FAN1 nuclease on DNA substrates harboring (CAG)2 extrahelical extrusions. (A) Time course of FAN1 nuclease activity on 3′-/Cy3/-labeled 40-mer linear DNAs harboring different extrahelical extrusions [(CAG)2 (blue), (CTG)2 (cyan), (AGCCTA) (red), (CTG)13 (pink), (CAG)13 (green), and homoduplex control (black)] was performed at 70 mM KCl. Data are mean of at least three independent experiments. Error bars represent SD. (B) Schematic of the FAN1 nuclease assay on (CAG)2 extrusion harboring circular DNA under low or near-physiological ionic strength. The red and blue arrows indicate the location of the extrusion and the nick, respectively. (C) FAN1 cleavage of the C strand (brown) or the V strand (blue) of a 3′(CAG)2 DNA substrate was determined at different ionic strengths. Reaction products were digested with ScaI, resolved on 1% alkaline agarose gels, followed by indirect end labeling with 5′ digoxigenin (/5DigN/) labeled probe (Fwd1947) to visualize C strand (brown) or Rev1975 probe to visualize V strand (blue) (SI Appendix, Materials and Methods and Fig. S1B). Values are mean of n ≥ 3 independent experiments (± SD). (D) 3′(CAG)2 (lanes 1 to 9) or 3′ control homoduplex (lanes 10 to 12) were incubated in the presence or absence of FAN1 (or FAN1 D960A), PCNA, and RFC, as indicated. The reactions were performed at 125 mM KCl in the presence of ATP (except lane 9). Products were digested with ScaI, resolved on 1% alkaline agarose gels, followed by indirect end labeling with Fwd1947 probe, Rev1975 probe (E), or Fwd2020 probe (F). M- marker; mr78 4xLacO plasmid was digested with BglII (to indicate the location of (CAG)2 extrusion- red arrowhead) or BbvCI (to indicate the location of the nick- blue arrowhead). (G) Products of the reaction were also digested with PspFI and resolved on 10% polyacrylamide gels containing 8 M urea, followed by indirect end labeling with Fwd3028 probe. The size marker was generated by digestion of mr77 4xLacO with AlwNI, XbaI, or AatII with the distance from the nick indicated on the side. The mobility of the full-length-labeled DNA segment is indicated by asterisk. See also SI Appendix, Fig. S1.