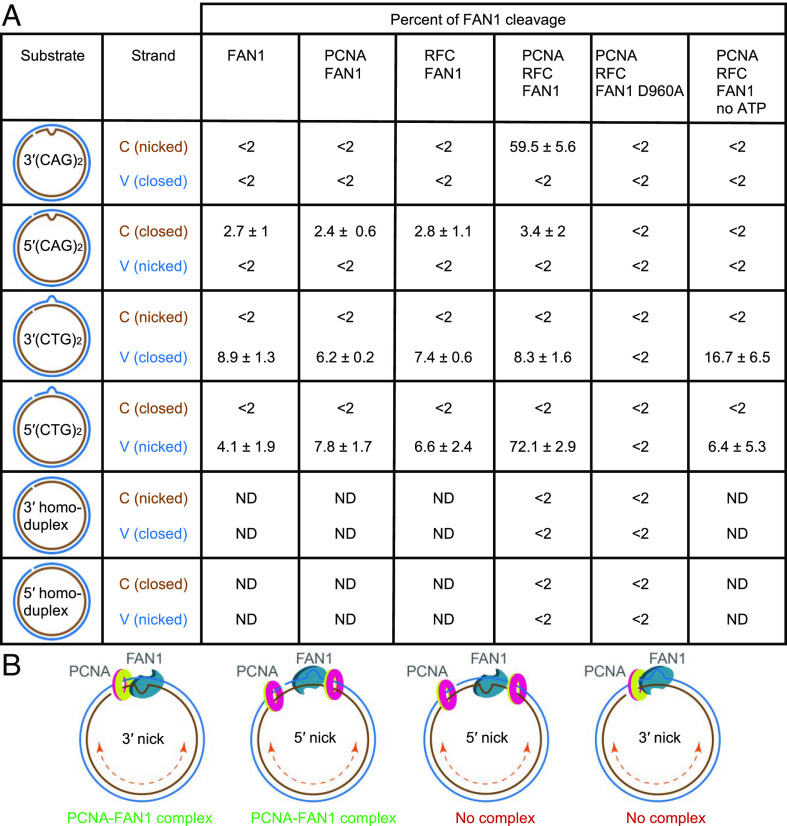
Fig. 2.
Strand directionality of PCNA- and RFC-dependent activation of the FAN1 nuclease. (A) FAN1 nuclease cleavage of C strand (brown) or V strand (blue) of extrusion-containing circular DNA substrates or homoduplex controls (as shown) in the presence of the indicated proteins was determined using indirect end labeling with Fwd1947 or Rev1975 probes, respectively, and quantified as described (SI Appendix, Materials and Methods). Data are mean of at least three independent experiments ± SD, except for 3′(CTG)2 DNA, presented data is an average of two independent experiments with range observed. Representative images are shown in Fig. 1 and SI Appendix, Fig. S2. ND, not determined. (B) Proposed mechanism for FAN1 interaction with DNA-loaded PCNA. On the 3′(CAG)2 or 5′(CTG)2 DNA substrates (strand break and the extrusion are on the same DNA strand), PCNA and FAN1 can form a complex leading to activation of FAN1 nuclease. On the 3′(CTG)2 or 5′(CAG)2 substrates (strand break and the extrusion are on the opposite DNA strands), PCNA and FAN1 complex does not form, and no DNA cleavage by FAN1 is observed. See also SI Appendix, Fig. S2.